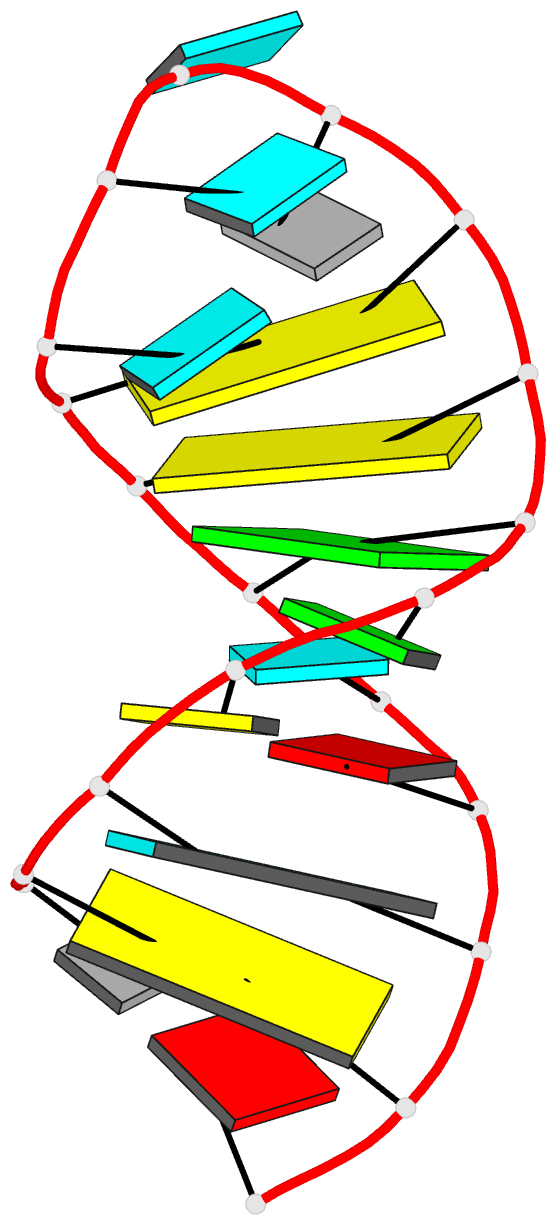
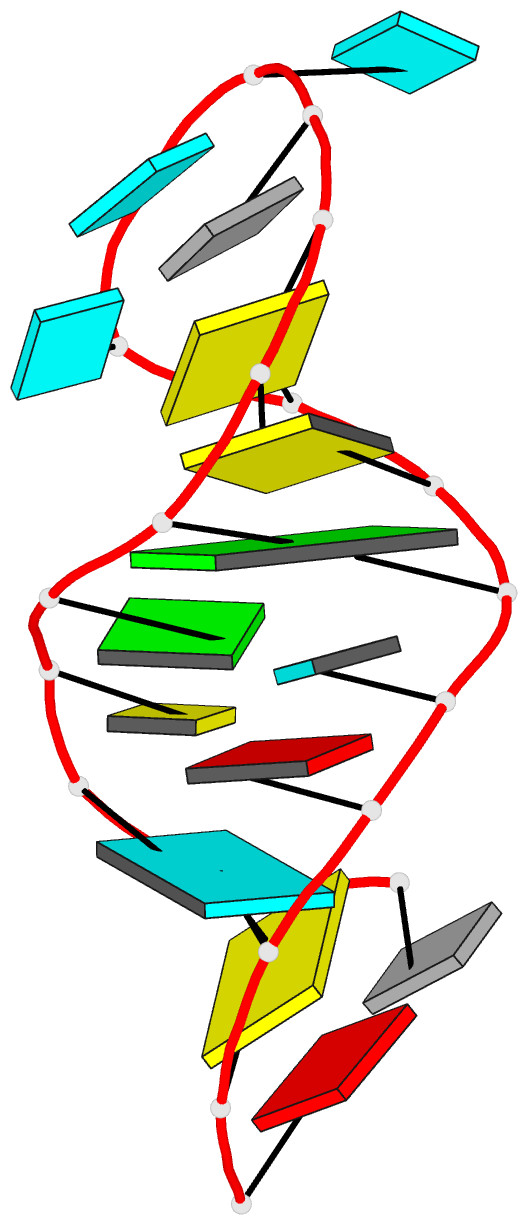
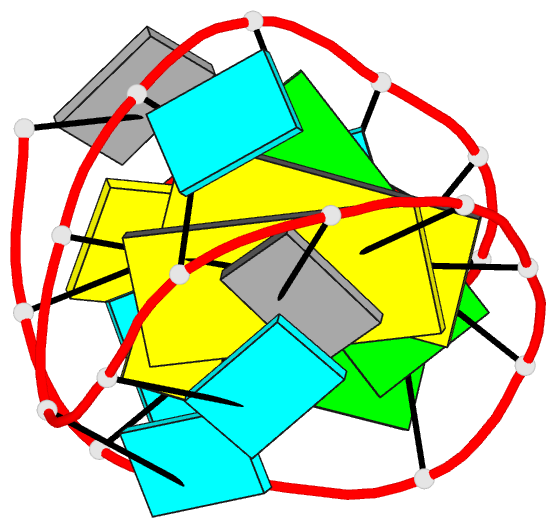
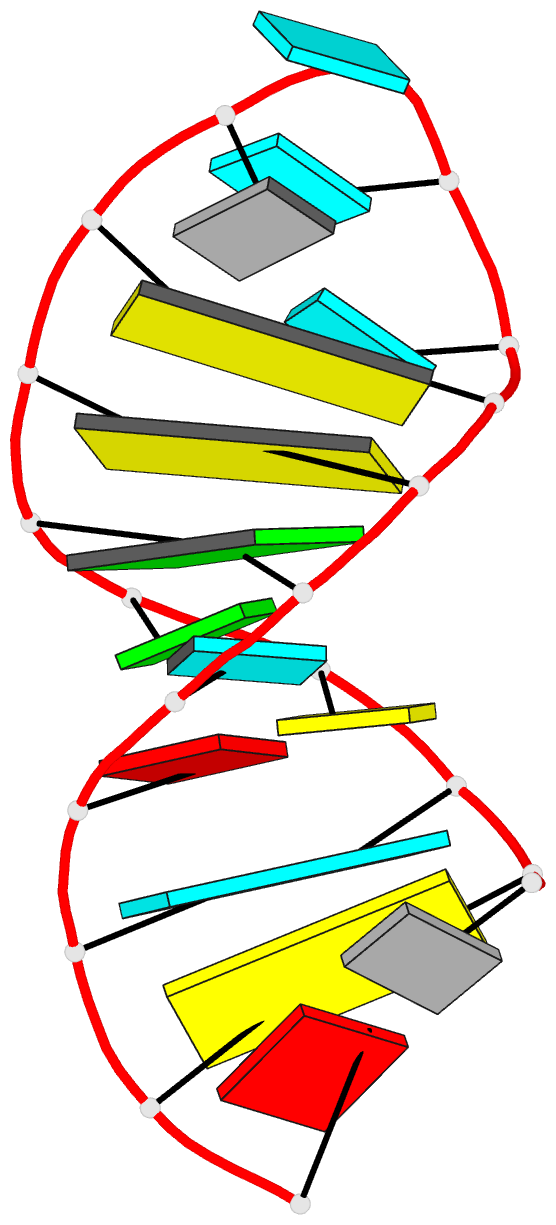
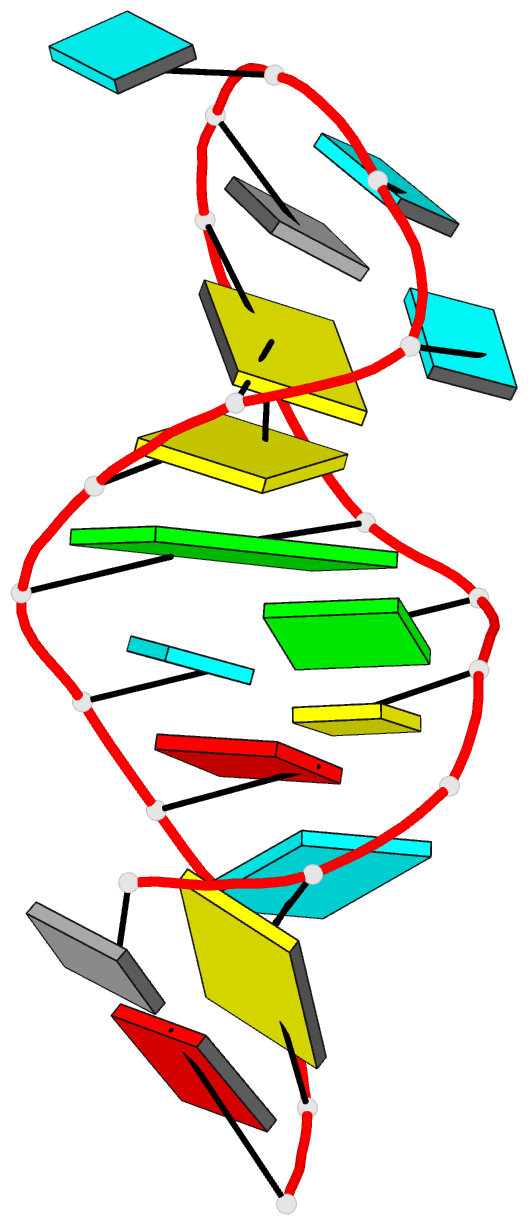
Summary information and primary citation
- PDB-id
- 2o32; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Solution structure of u2 snrna stem i from human, containing modified nucleotides
- Reference
- Sashital DG, Venditti V, Angers CG, Cornilescu G, Butcher SE (2007): "Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human." Rna, 13, 328-338. doi: 10.1261/rna.418407.
- Abstract
- The spliceosome is a dynamic ribonucleoprotein complex responsible for the removal of intron sequences from pre-messenger RNA. The highly conserved 5' end of the U2 small nuclear RNA (snRNA) makes key base-pairing interactions with the intron branch point sequence and U6 snRNA. U2 stem I, a stem-loop located in the 5' region of U2, has been implicated in spliceosome assembly and may modulate the folding of the U2 and U6 snRNAs in the spliceosome active site. Here we present the NMR structures of U2 stem I from human and Saccharomyces cerevisiae. These sequences represent the two major classes of U2 stem I, distinguished by the identity of tandem wobble pairs (UU/UU in yeast and CA/GU in human) and the presence of post-transcriptional modifications (four 2'-O-methyl groups and two pseudouracils in human). The structures reveal that the UU/UU and CA/GU tandem wobble pairs are nearly isosteric. The tandem wobble pairs separate two thermodynamically distinct regions of Watson-Crick base pairs, with the modified nucleotides in human stem I conferring a significant increase in stability. We hypothesize that the separate thermodynamic stabilities of U2 stem I exist to allow the structure to transition through different folded conformations during spliceosome assembly and catalysis.