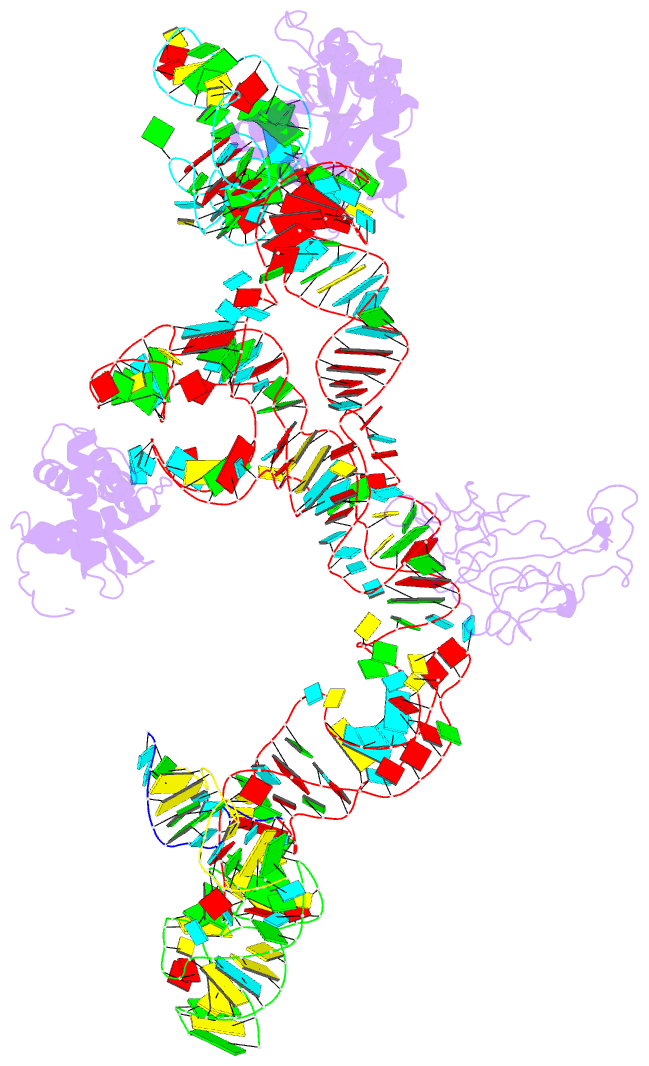
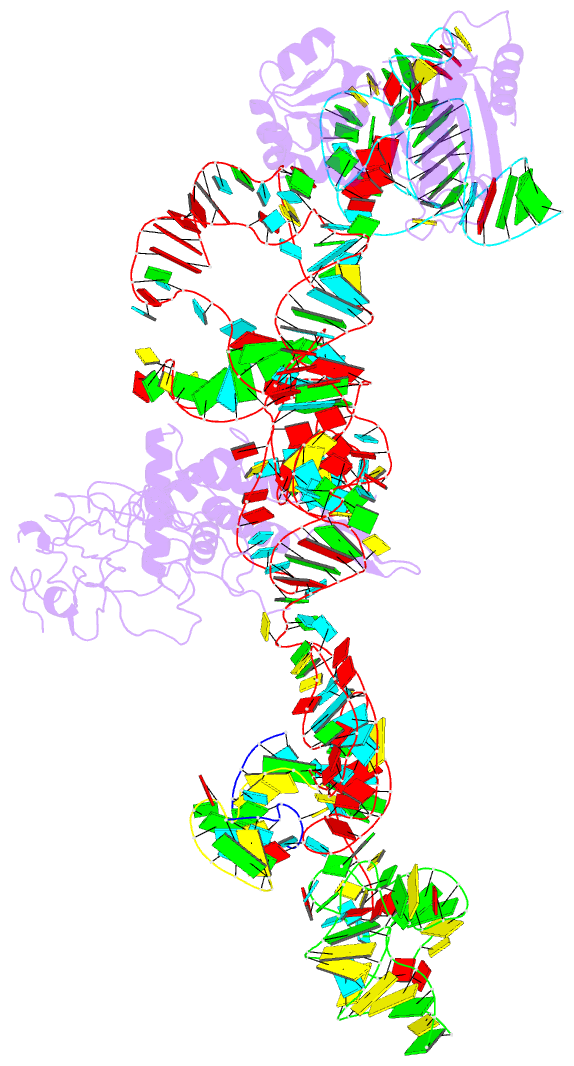
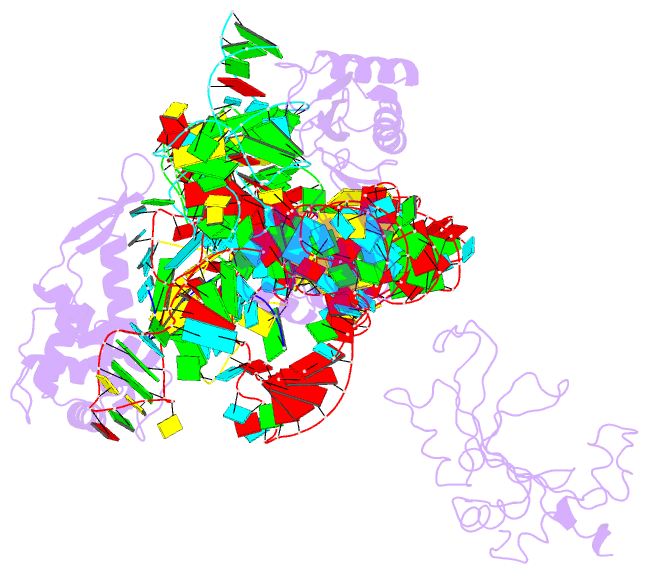
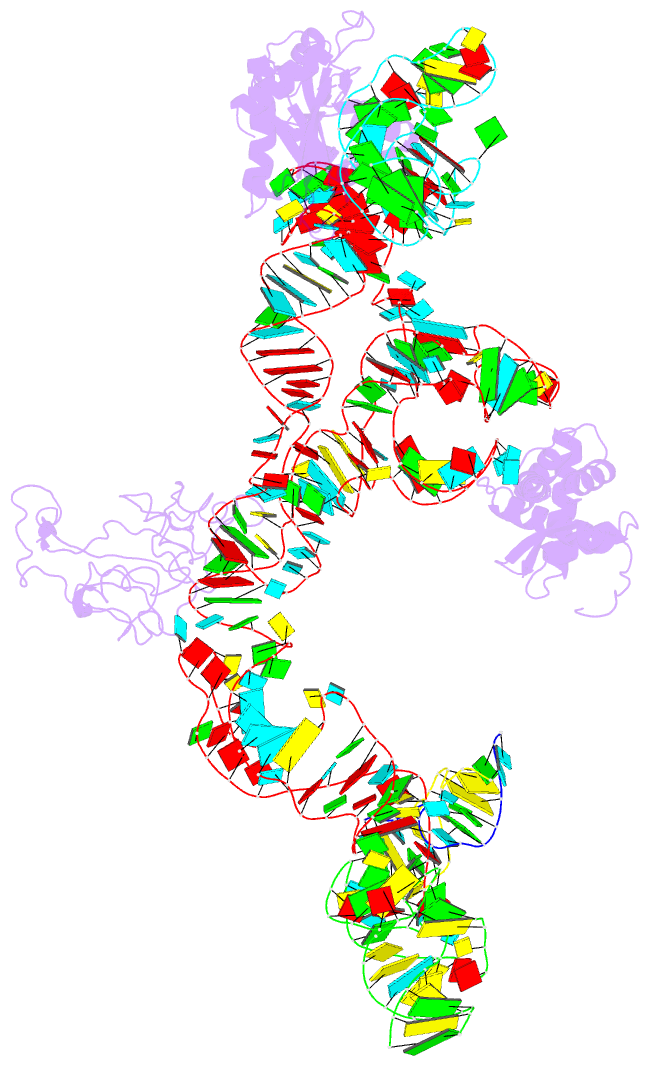
Summary information and primary citation
- PDB-id
- 2noq; DSSR-derived features in text and JSON formats
- Class
- ribosome
- Method
- cryo-EM (7.3 Å)
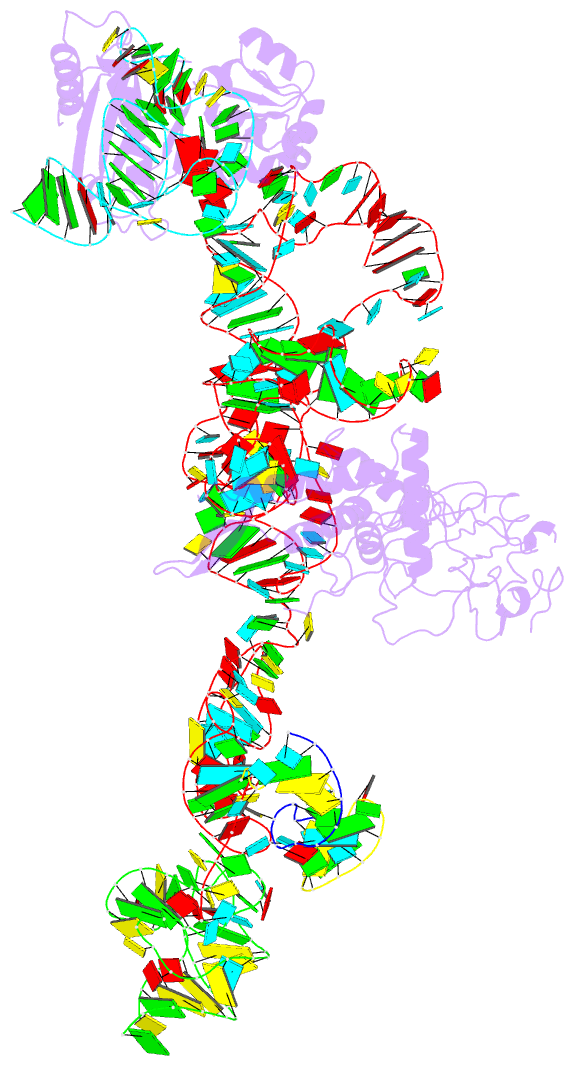
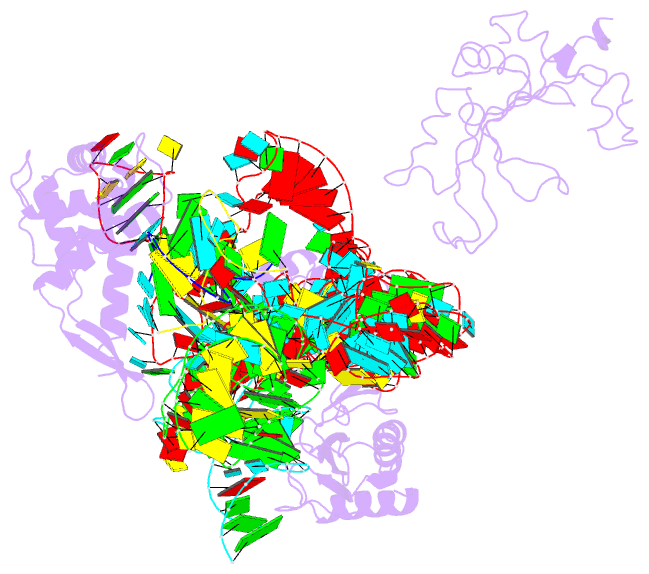
- Summary
- Structure of ribosome-bound cricket paralysis virus ires RNA
- Reference
- Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM (2006): "Structure of the ribosome-bound cricket paralysis virus IRES RNA." Nat.Struct.Mol.Biol., 13, 1092-1096. doi: 10.1038/nsmb1177.
- Abstract
- Internal ribosome entry sites (IRESs) facilitate an alternative, end-independent pathway of translation initiation. A particular family of dicistroviral IRESs can assemble elongation-competent 80S ribosomal complexes in the absence of canonical initiation factors and initiator transfer RNA. We present here a cryo-EM reconstruction of a dicistroviral IRES bound to the 80S ribosome. The resolution of the cryo-EM reconstruction, in the subnanometer range, allowed the molecular structure of the complete IRES in its active, ribosome-bound state to be solved. The structure, harboring three pseudoknot-containing domains, each with a specific functional role, shows how defined elements of the IRES emerge from a compactly folded core and interact with the key ribosomal components that form the A, P and E sites, where tRNAs normally bind. Our results exemplify the molecular strategy for recruitment of an IRES and reveal the dynamic features necessary for internal initiation.