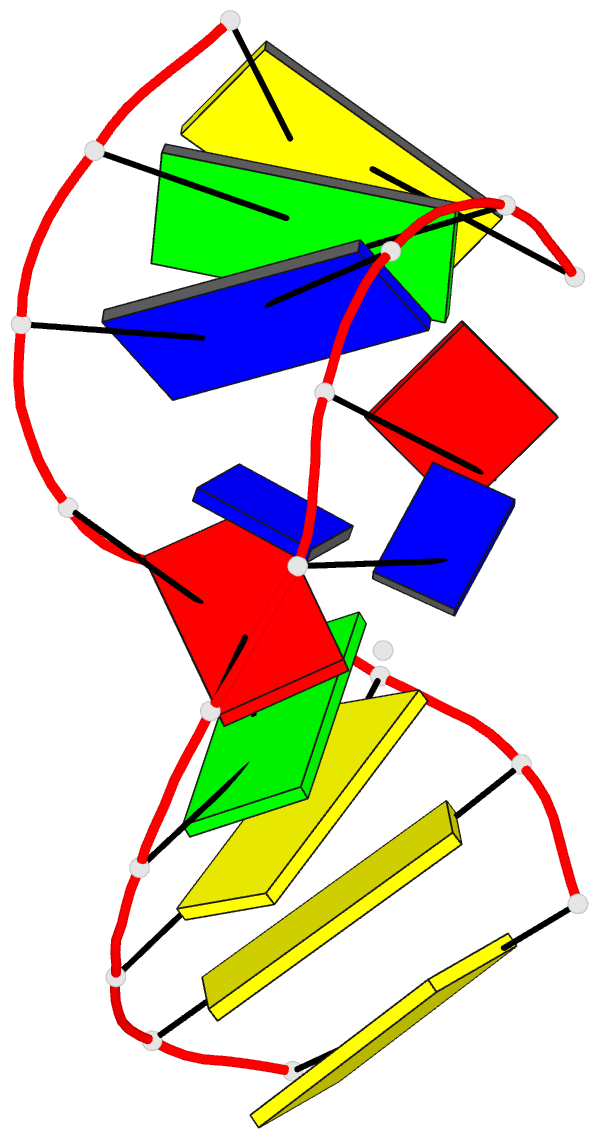
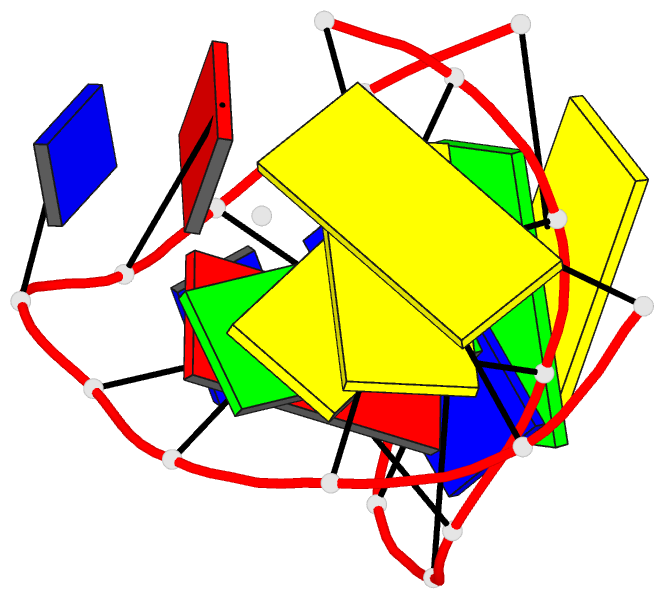
Summary information and primary citation
- PDB-id
- 2neo; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- NMR
- Summary
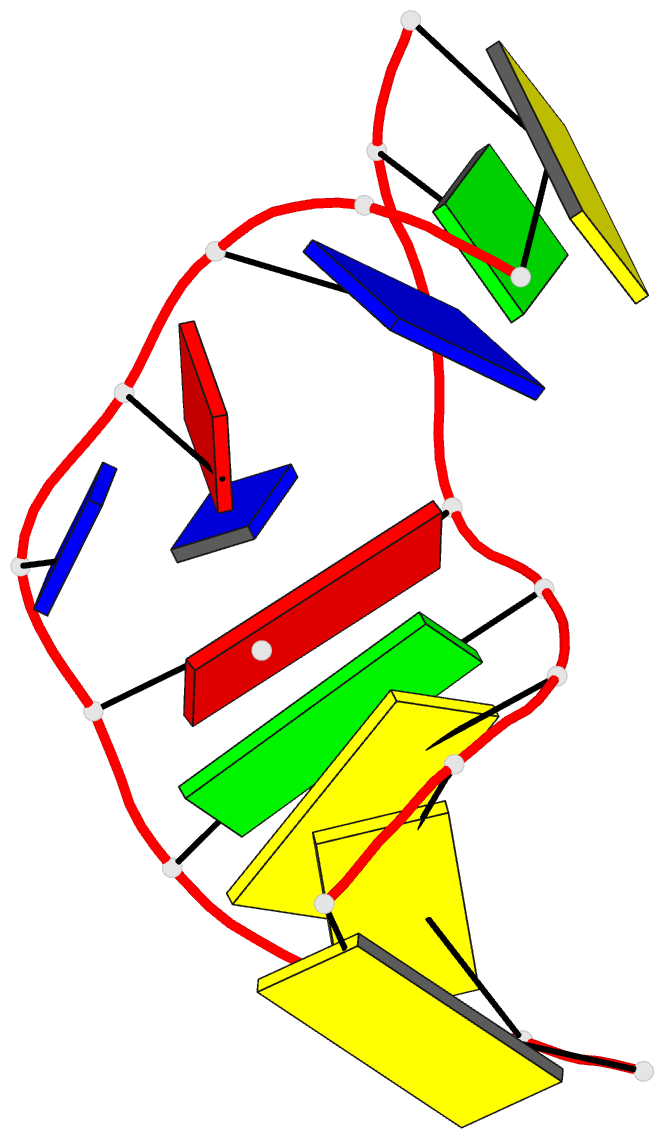
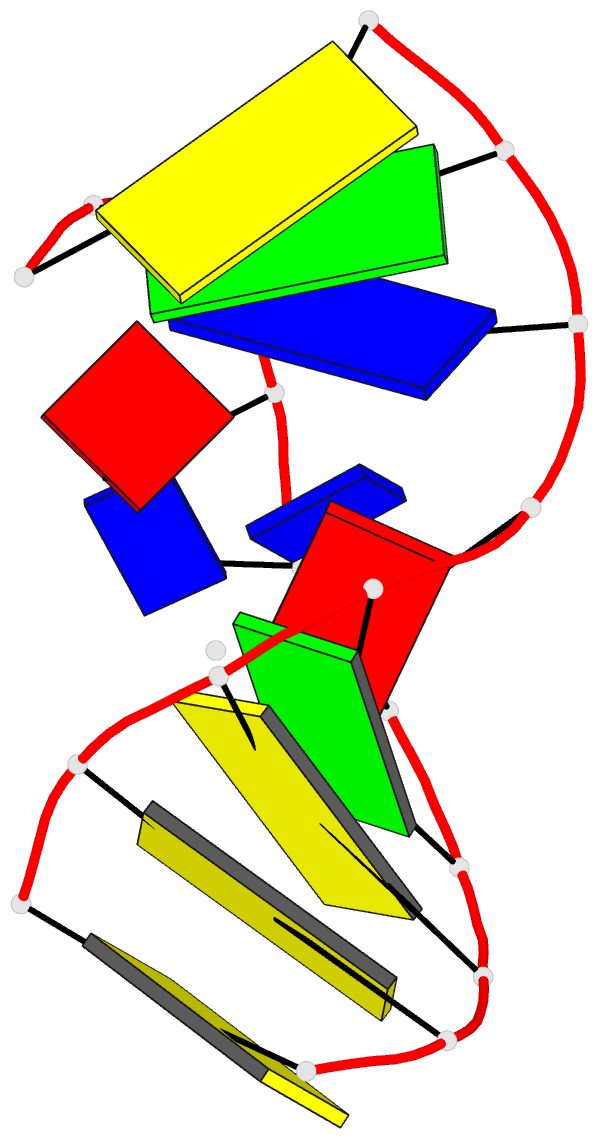
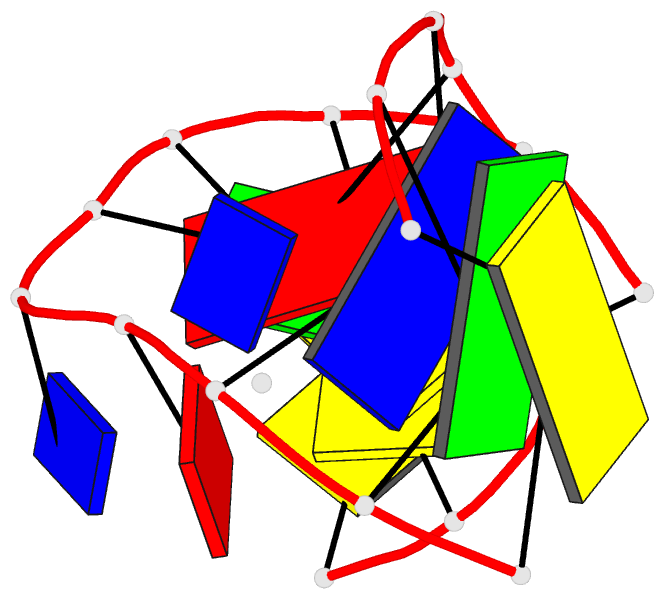
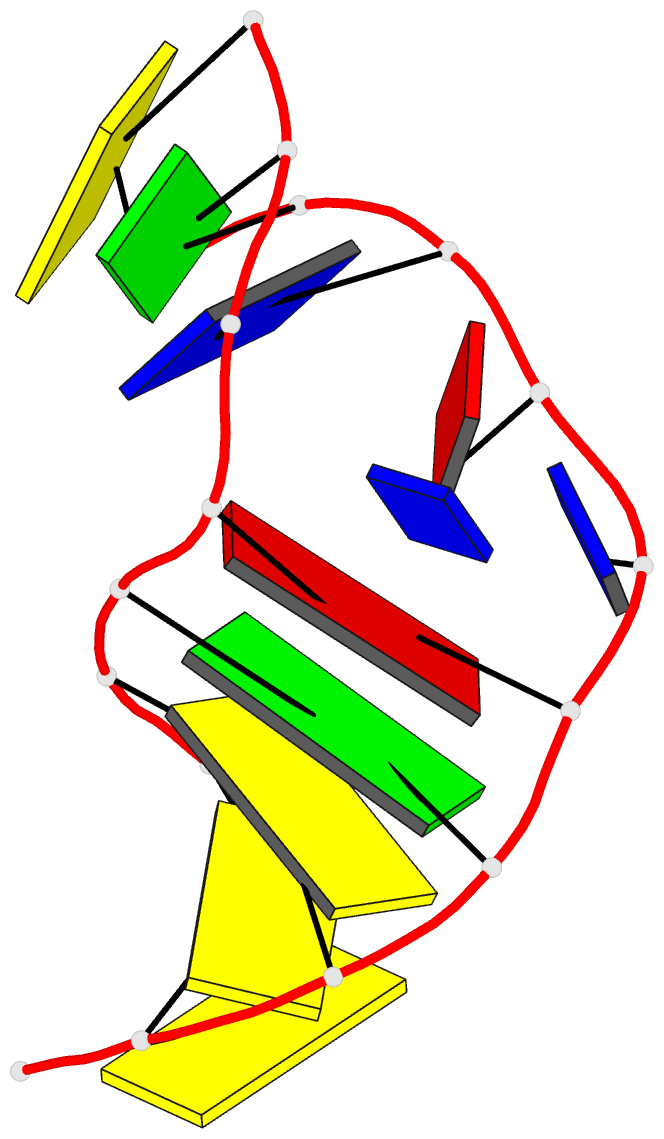
- Solution NMR structure of a two-base DNA bulge complexed with an enediyne cleaving analog, 11 structures
- Reference
- Stassinopoulos A, Ji J, Gao X, Goldberg IH (1996): "Solution structure of a two-base DNA bulge complexed with an enediyne cleaving analog." Science, 272, 1943-1946.
- Abstract
- Nucleic acid bulges have been implicated in a number of biological processes and are specific cleavage targets for the enediyne antitumor antibiotic neocarzinostatin chromophore in a base-catalyzed, radical-mediated reaction. The solution structure of the complex between an analog of the bulge-specific cleaving species and an oligodeoxynucleotide containing a two-base bulge was elucidated by nuclear magnetic resonance. An unusual binding mode involves major groove recognition by the drug carbohydrate unit and tight fitting of the wedge-shaped drug in the triangular prism pocket formed by the two looped-out bulge bases and the neighboring base pairs. The two drug rings mimic helical DNA bases, complementing the bent DNA structure. The putative abstracting drug radical is 2.2 +/- 0.1 angstroms from the pro-S H5' of the target bulge nucleotide. This structure clarifies the mechanism of bulge recognition and cleavage by a drug and provides insight into the design of bulge-specific nucleic acid binding molecules.