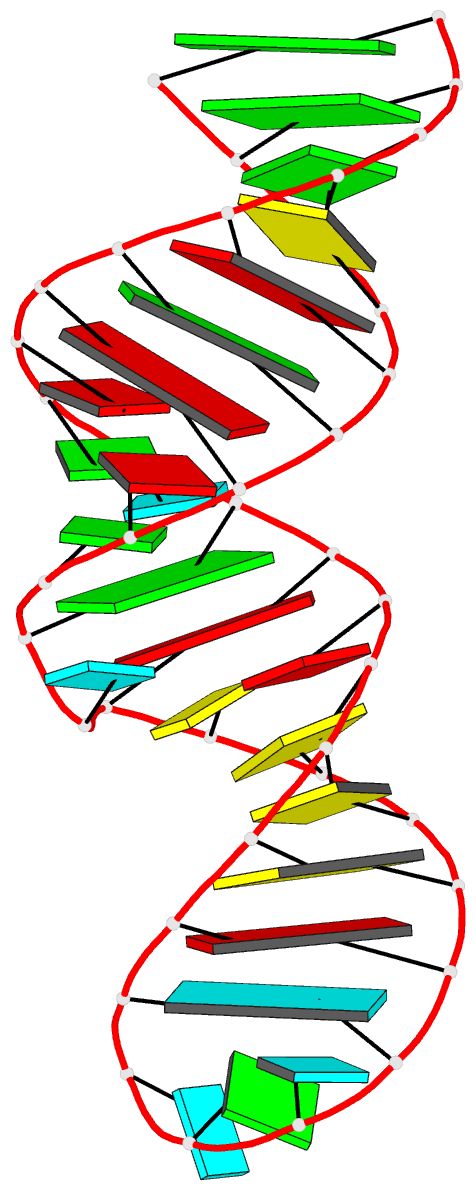
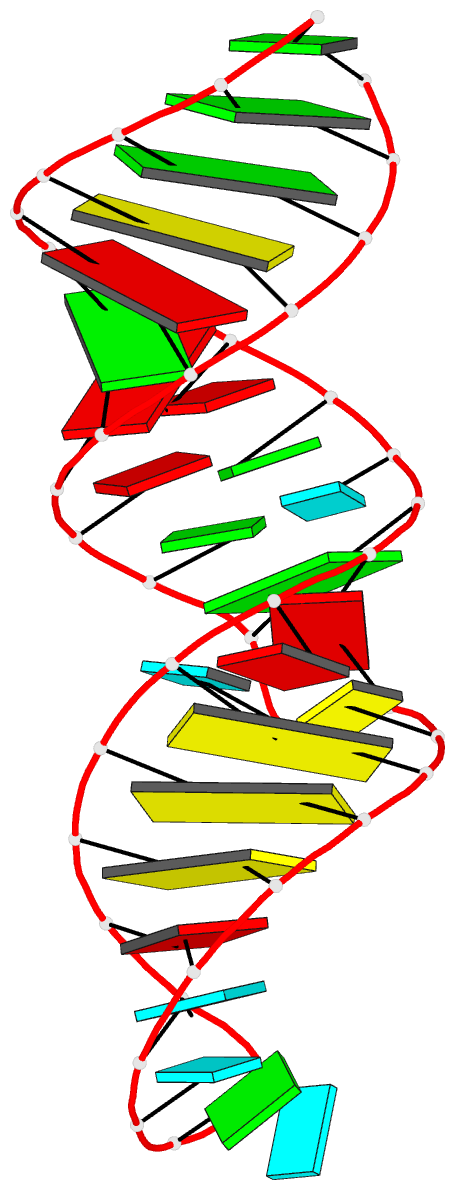
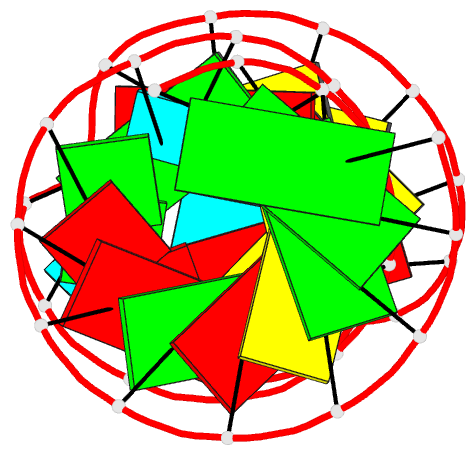
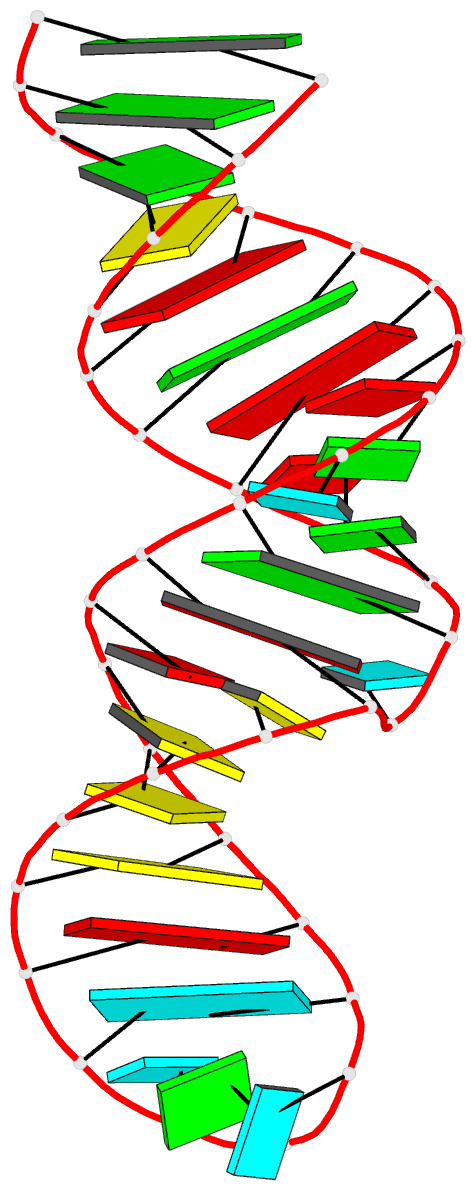
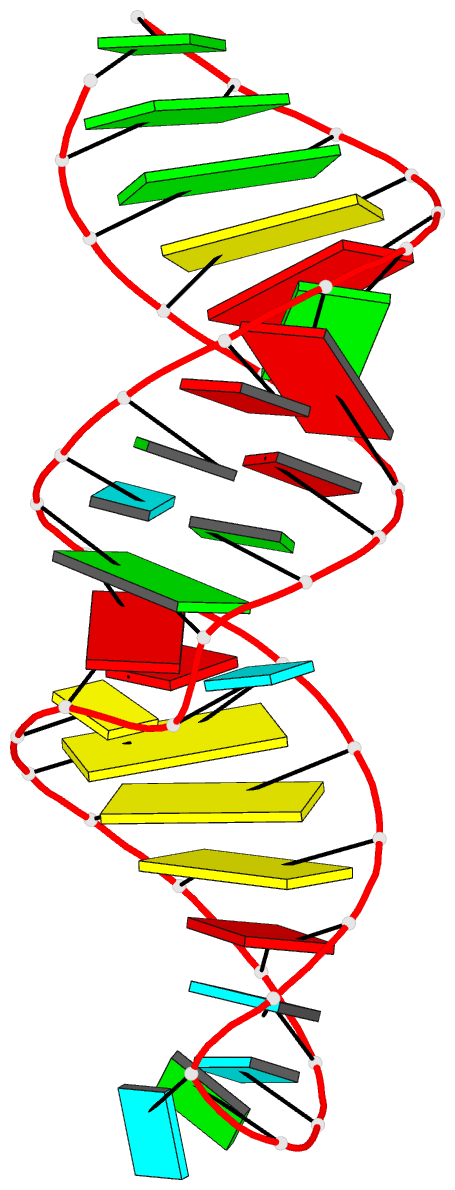
Summary information and primary citation
- PDB-id
- 2nby; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Solution structure of the j domain of emcv ires
- Reference
- Imai S, Kumar P, Hellen CU, D'Souza VM, Wagner G (2016): "An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation." Nat. Struct. Mol. Biol., 23, 859-864. doi: 10.1038/nsmb.3280.
- Abstract
- Many viruses bypass canonical cap-dependent translation in host cells by using internal ribosomal entry sites (IRESs) in their transcripts; IRESs hijack initiation factors for the assembly of initiation complexes. However, it is currently unknown how IRES RNAs recognize initiation factors that have no endogenous RNA binding partners; in a prominent example, the IRES of encephalomyocarditis virus (EMCV) interacts with the HEAT-1 domain of eukaryotic initiation factor 4G (eIF4G). Here we report the solution structure of the J-K region of this IRES and show that its stems are precisely organized to position protein-recognition bulges. This multisite interaction mechanism operates on an all-or-nothing principle in which all domains are required. This preorganization is accomplished by an 'adjuster module': a pentaloop motif that acts as a dual-sided docking station for base-pair receptors. Because subtle changes in the orientation abrogate protein capture, our study highlights how a viral RNA acquires affinity for a target protein.