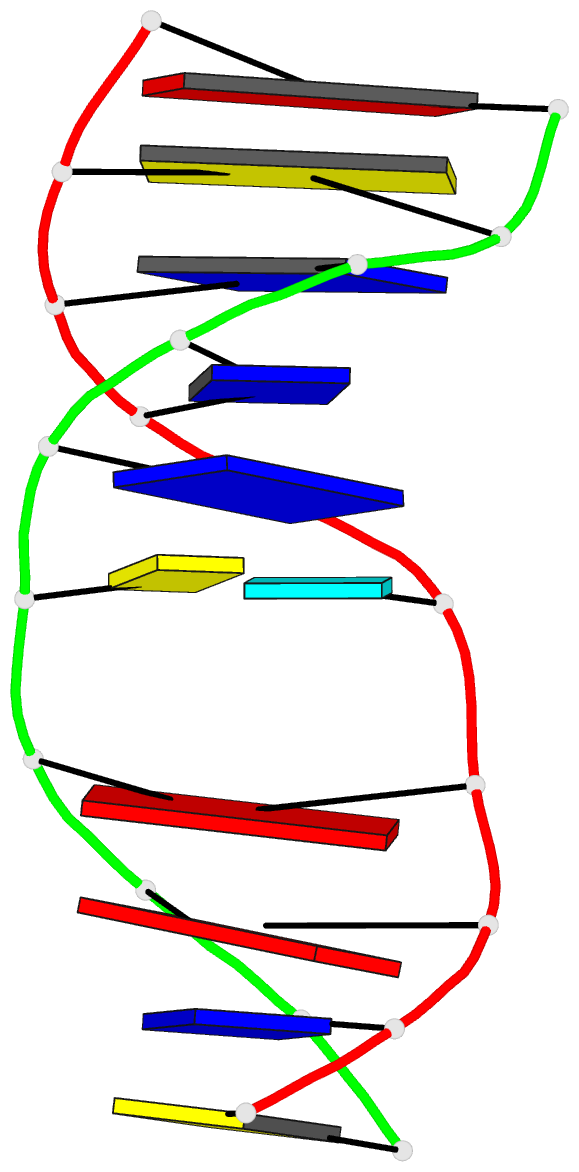
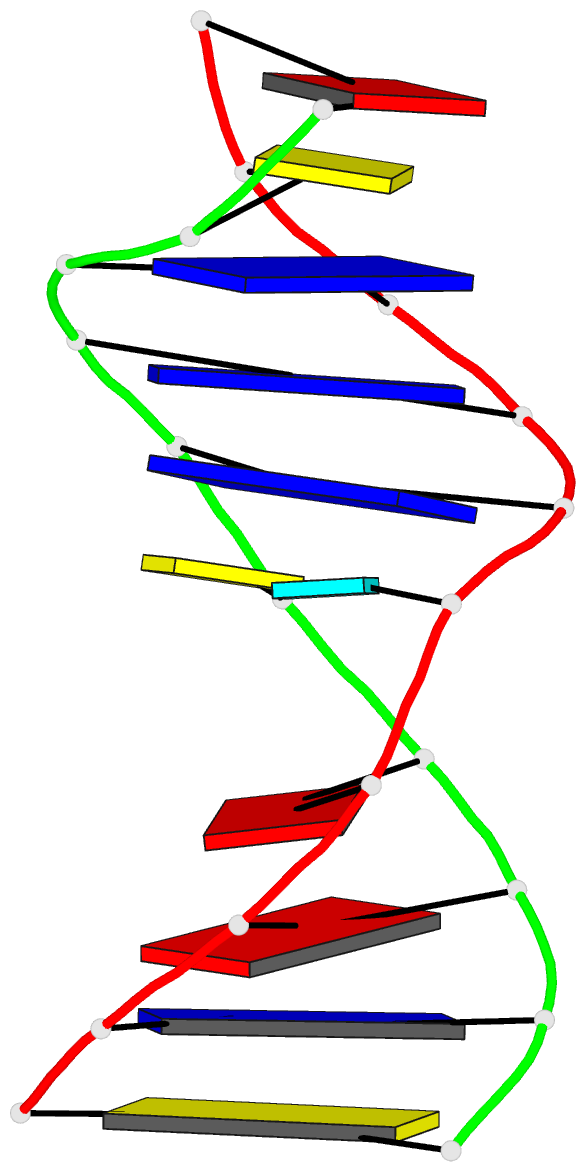
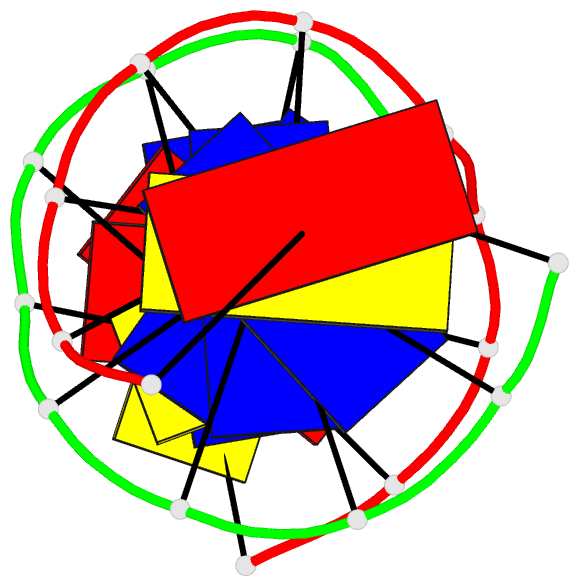
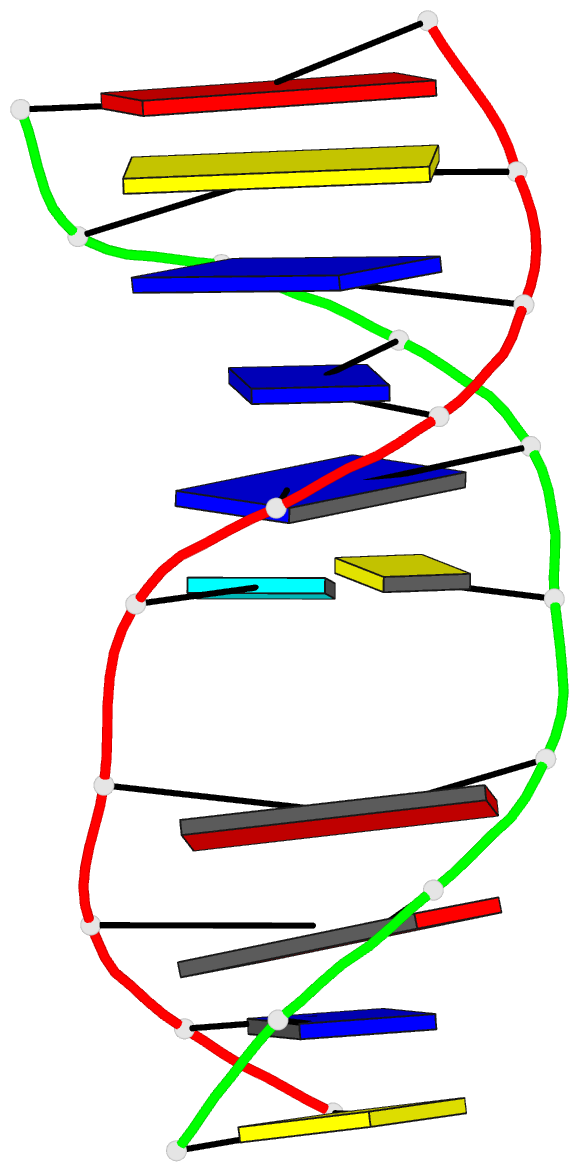
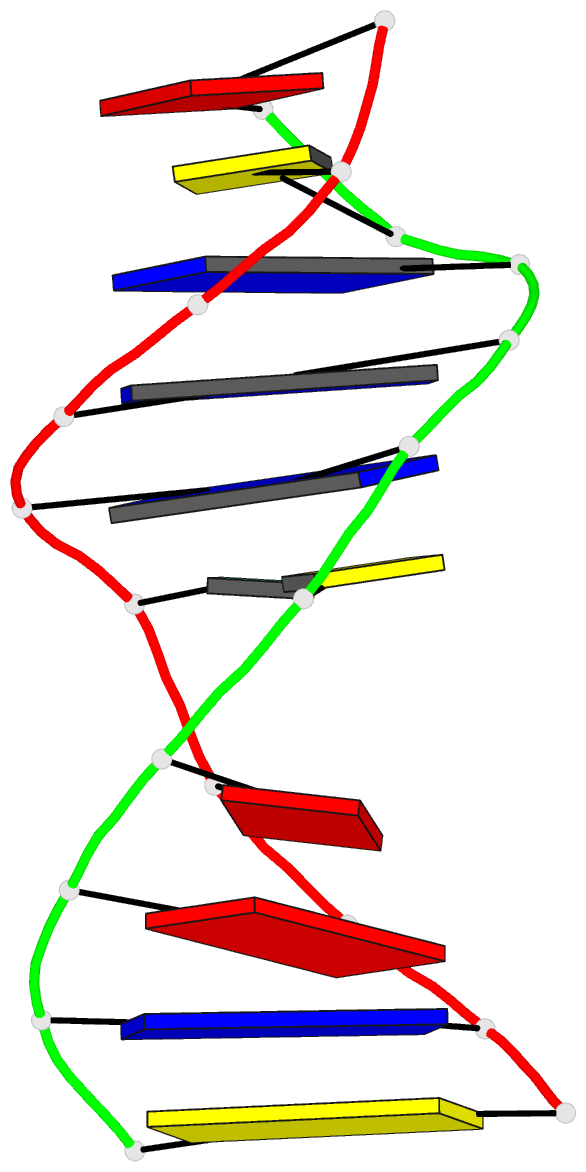
Summary information and primary citation
- PDB-id
- 2mmq; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of agt fapy modified duplex
- Reference
- Li L, Brown KL, Ma R, Stone MP (2015): "DNA Sequence Modulates Geometrical Isomerism of the trans-8,9-Dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy Aflatoxin B1 Adduct." Chem.Res.Toxicol., 28, 225-237. doi: 10.1021/tx5003832.
- Abstract
- Aflatoxin B1 (AFB1), a mycotoxin produced by Aspergillus flavus, is oxidized by cytochrome P450 enzymes to aflatoxin B1-8,9-epoxide, which alkylates DNA at N7-dG. Under basic conditions, this N7-dG adduct rearranges to yield the trans-8,9-dihydro-8-(2,6-diamino-4-oxo-3,4-dihydropyrimid-5-yl-formamido)-9-hydroxy aflatoxin B1 (AFB1-FAPY) adduct. The AFB1-FAPY adduct exhibits geometrical isomerism involving the formamide moiety. NMR analyses of duplex oligodeoxynucleotides containing the 5'-XA-3', 5'-XC-3', 5'-XT-3', and 5'-XY-3' sequences (X = AFB1-FAPY; Y = 7-deaza-dG) demonstrate that the equilibrium between E and Z isomers is controlled by major groove hydrogen bonding interactions. Structural analysis of the adduct in the 5'-XA-3' sequence indicates the preference of the E isomer of the formamide group, attributed to formation of a hydrogen bond between the formyl oxygen and the N(6) exocyclic amino group of the 3'-neighbor adenine. While the 5'-XA-3' sequence exhibits the E isomer, the 5'-XC-3' sequence exhibits a 7:3 E:Z ratio at equilibrium at 283 K. The E isomer is favored by a hydrogen bond between the formyl oxygen and the N(4)-dC exocyclic amino group of the 3'-neighbor cytosine. The 5'-XT-3' and 5'-XY-3' sequences cannot form such a hydrogen bond between the formyl oxygen and the 3'-neighbor T or Y, respectively, and in these sequence contexts the Z isomer is favored. Additional equilibria between α and β anomers and the potential to exhibit atropisomers about the C5-N(5) bond do not depend upon sequence. In each of the four DNA sequences, the AFB1-FAPY adduct maintains the β deoxyribose configuration. Each of these four sequences feature the atropisomer of the AFB1 moiety that is intercalated above the 5'-face of the damaged guanine. This enforces the Ra axial conformation for the C5-N(5) bond.