Summary information and primary citation
- PDB-id
-
2lx1;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- NMR
- Summary
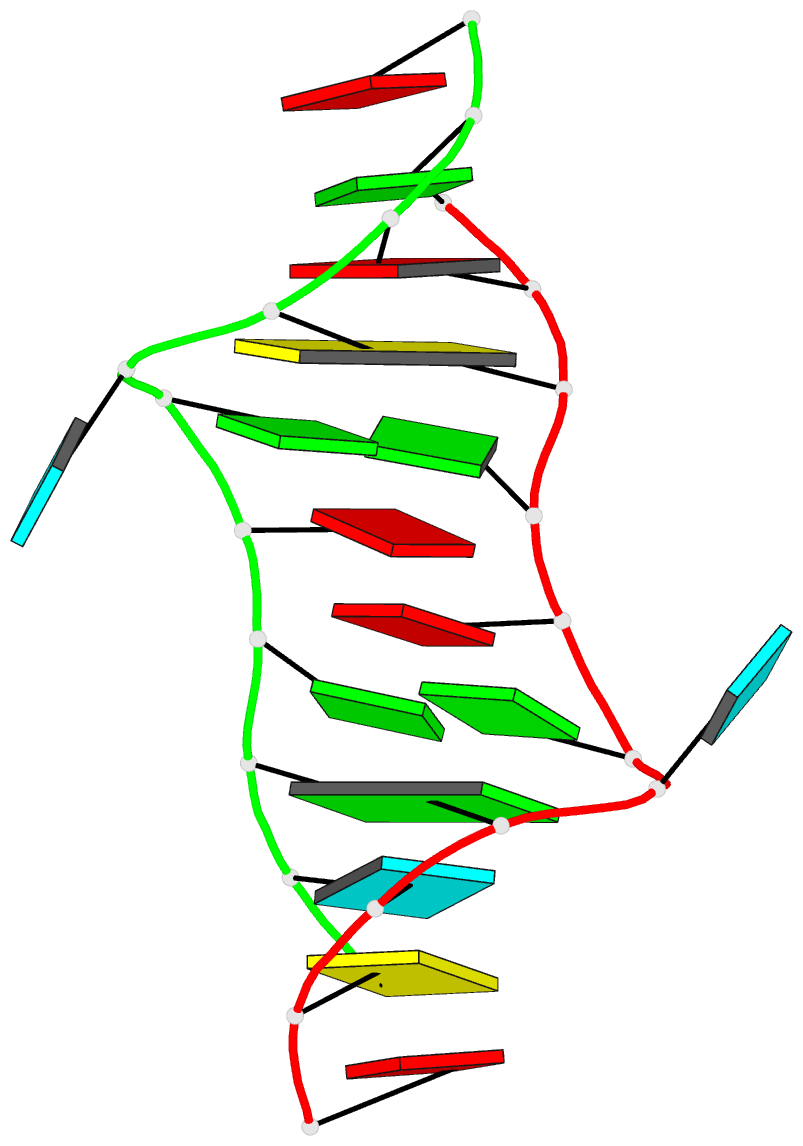
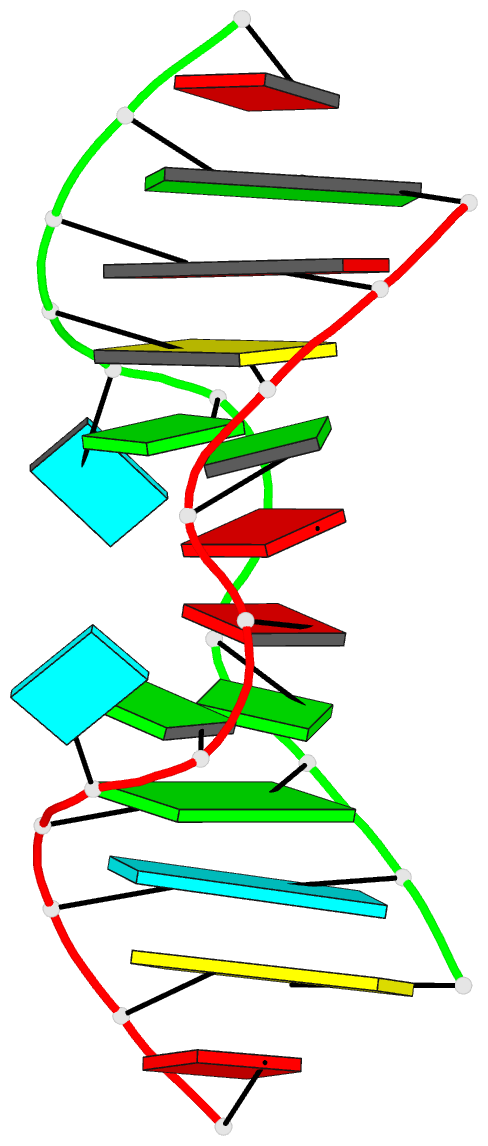
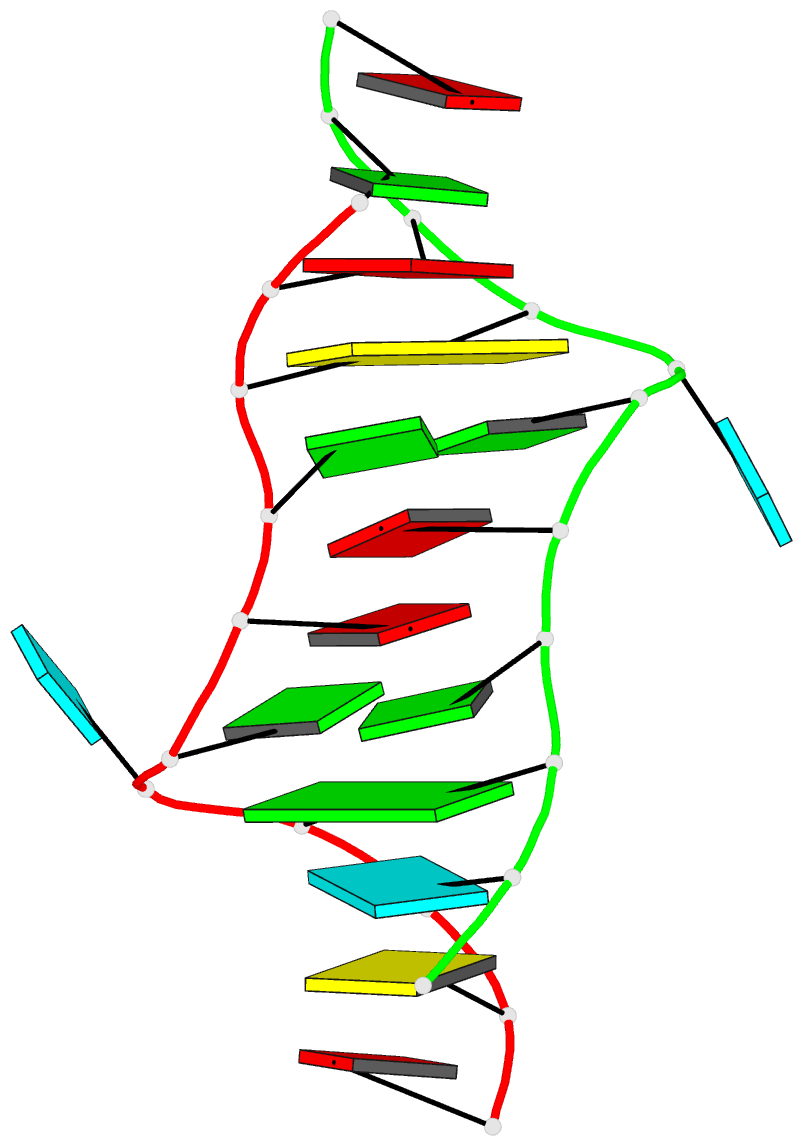
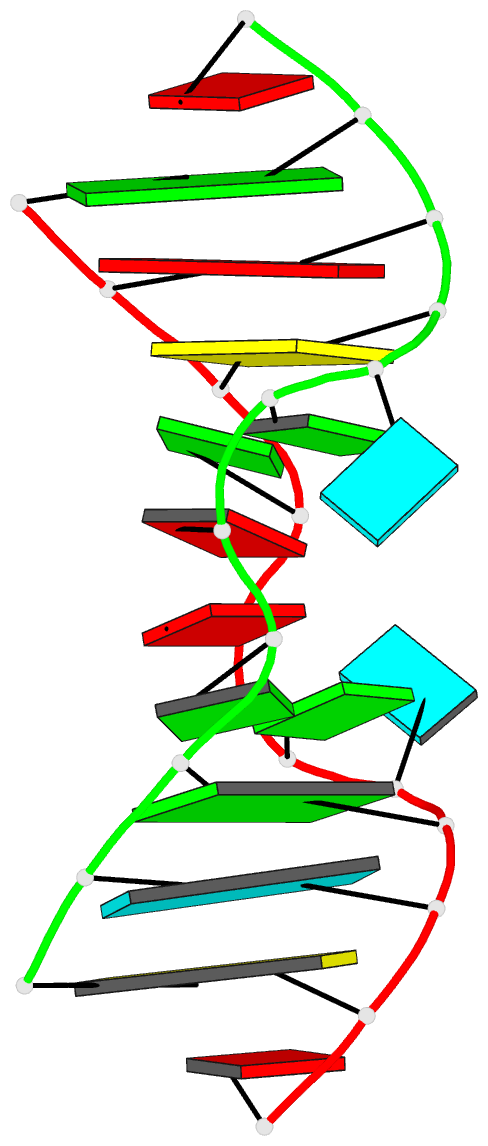
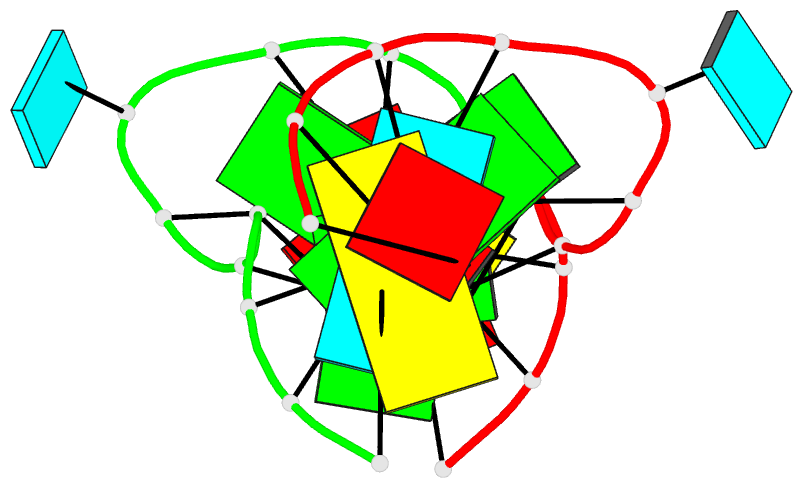
- Major conformation of the internal loop
5'gagu-3'ugag
- Reference
-
Kennedy SD, Kierzek R, Turner DH (2012): "Novel
conformation of an RNA structural switch."
Biochemistry, 51, 9257-9259.
doi: 10.1021/bi301372t.
- Abstract
- The RNA duplex, (5'GACGAGUGUCA)(2), has two
conformations in equilibrium. The nuclear magnetic
resonance solution structure reveals that the major
conformation of the loop, 5'GAGU/3'UGAG, is novel and
contains two unusual Watson-Crick/Hoogsteen GG pairs with G
residues in the syn conformation, two A residues stacked on
each other in the center of the helix with inverted sugars,
and two bulged-out U residues. The structure provides a
benchmark for testing approaches for predicting local RNA
structure and a sequence that allows the design of a unique
arrangement of functional groups and/or a conformational
switch into nucleic acids.