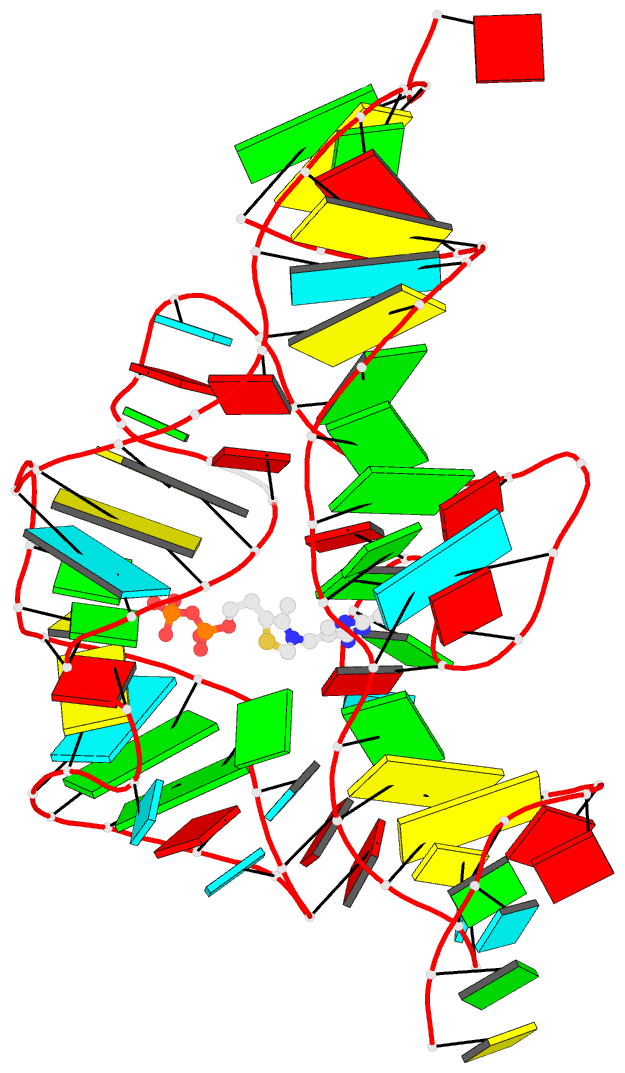
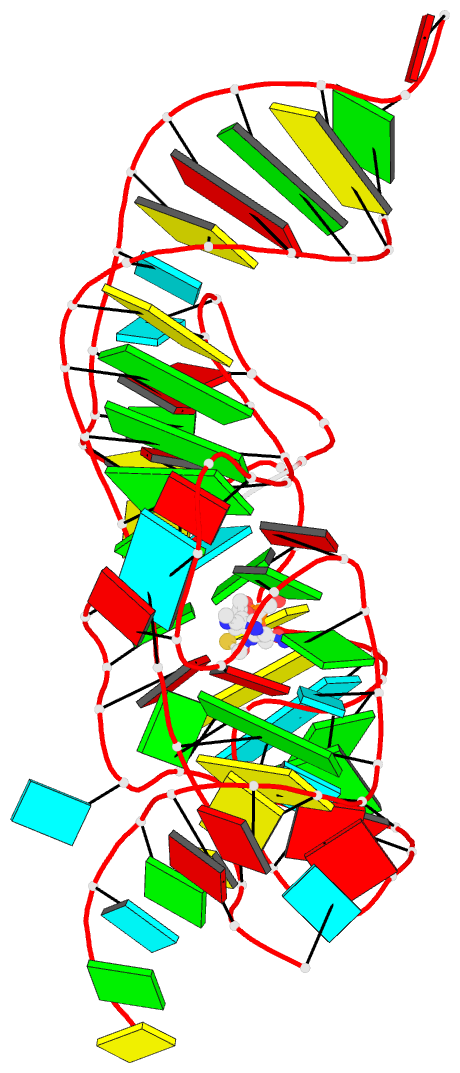
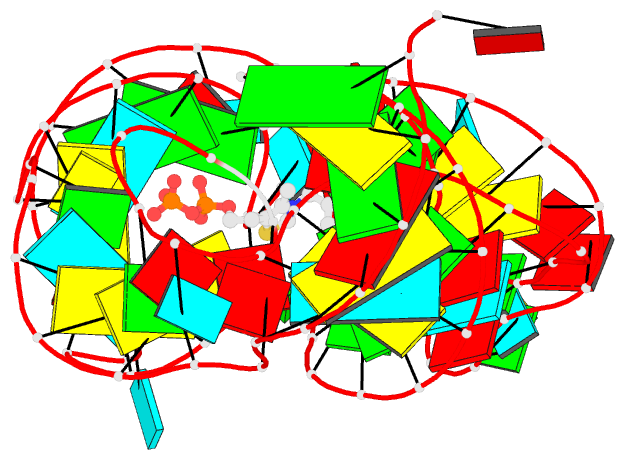
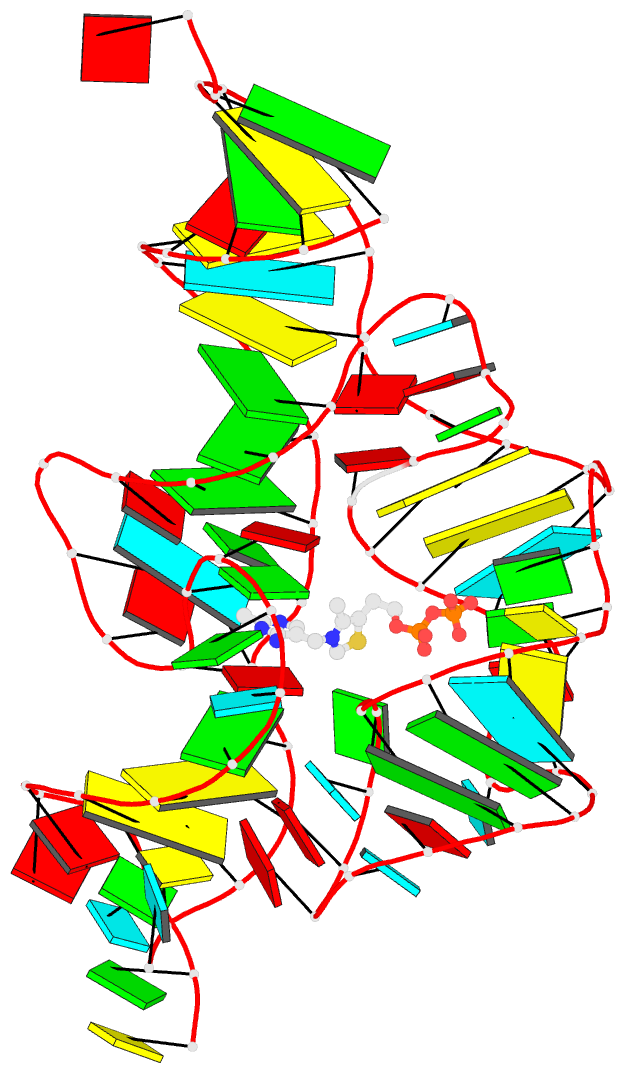
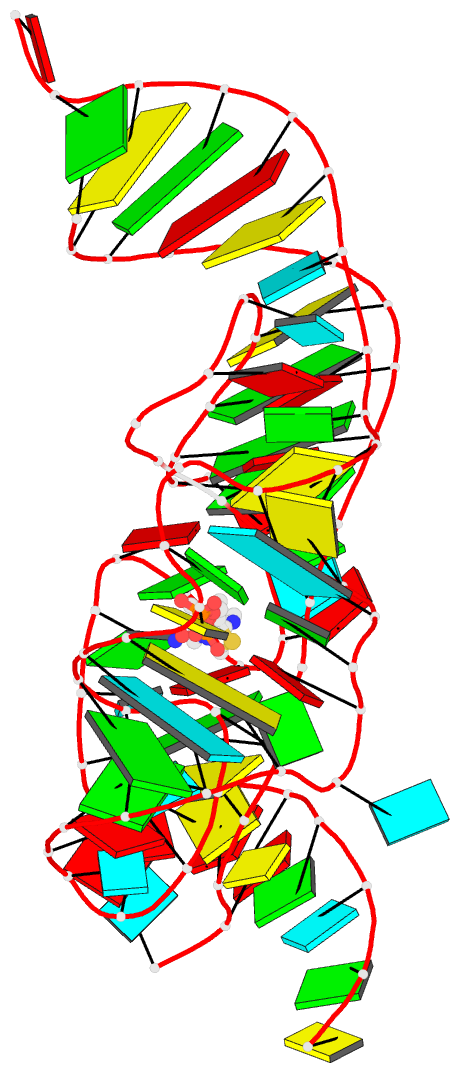
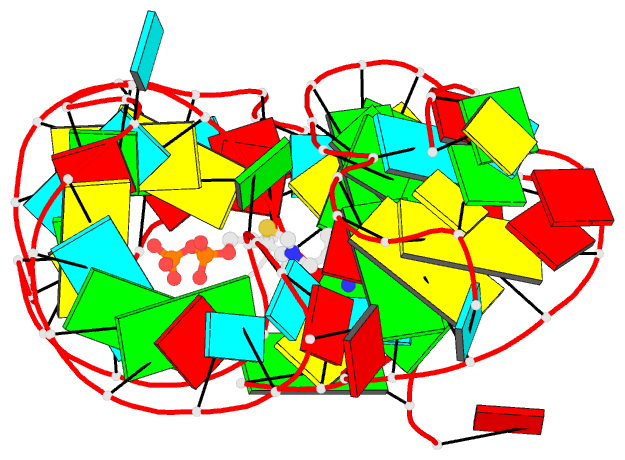
Summary information and primary citation
- PDB-id
-
2hoj;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- X-ray (2.5 Å)
- Summary
- Crystal structure of an e. coli thi-box riboswitch
bound to thiamine pyrophosphate, manganese ions
- Reference
-
Edwards TE, Ferre-D'Amare AR (2006): "Crystal
Structures of the Thi-Box Riboswitch Bound to Thiamine
Pyrophosphate Analogs Reveal Adaptive RNA-Small Molecule
Recognition." Structure,
14, 1459-1468. doi: 10.1016/j.str.2006.07.008.
- Abstract
- Riboswitches are noncoding mRNA elements that bind
small-molecule metabolites with high affinity and
specificity, and they regulate the expression of associated
genes. The thi-box riboswitch can exhibit a 1000-fold
higher affinity for thiamine pyrophosphate over closely
related noncognate compounds such as thiamine
monophosphate. To understand the chemical basis of thi-box
pyrophosphate specificity, we have determined crystal
structures of an E. coli thi-box bound to thiamine
pyrophosphate, thiamine monophosphate, and the structural
analogs benfotiamine and pyrithiamine. When bound to
monophosphorylated compounds, the RNA elements that
recognize the thiamine and phosphate moieties of the ligand
move closer together. This allows the riboswitch to
recognize the monophosphate in a manner similar to how it
recognizes the beta-phosphate of thiamine pyrophosphate. In
the pyrithiamine complex, the pyrophosphate binding site is
largely unstructured. These results show how the riboswitch
can bind to various metabolites, and why the thi-box
preferentially binds thiamine pyrophosphate.