Summary information and primary citation
- PDB-id
- 2h0z; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (2.7 Å)
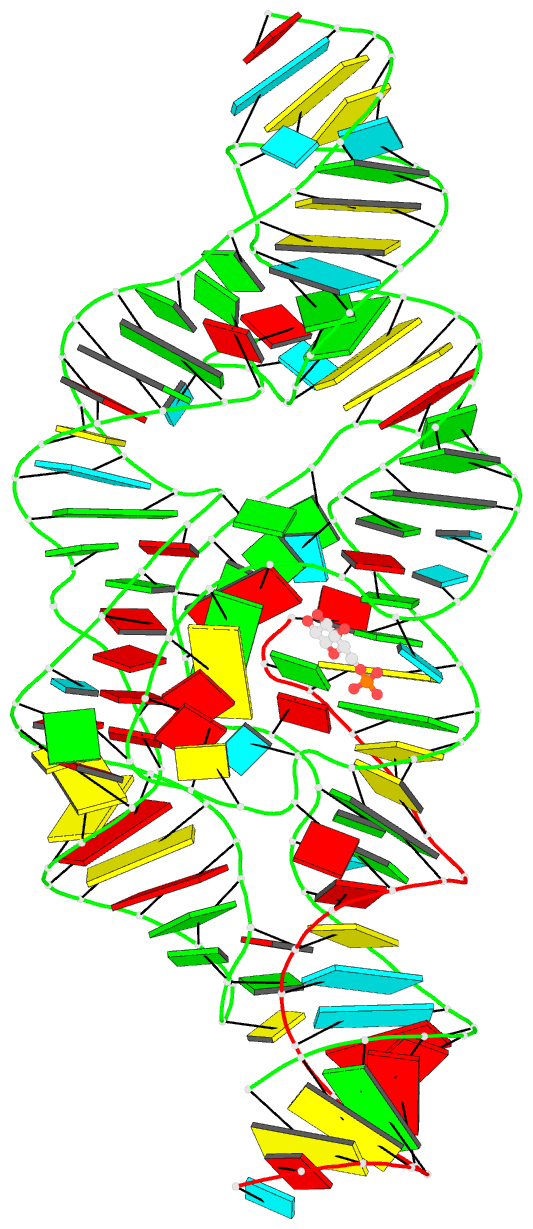
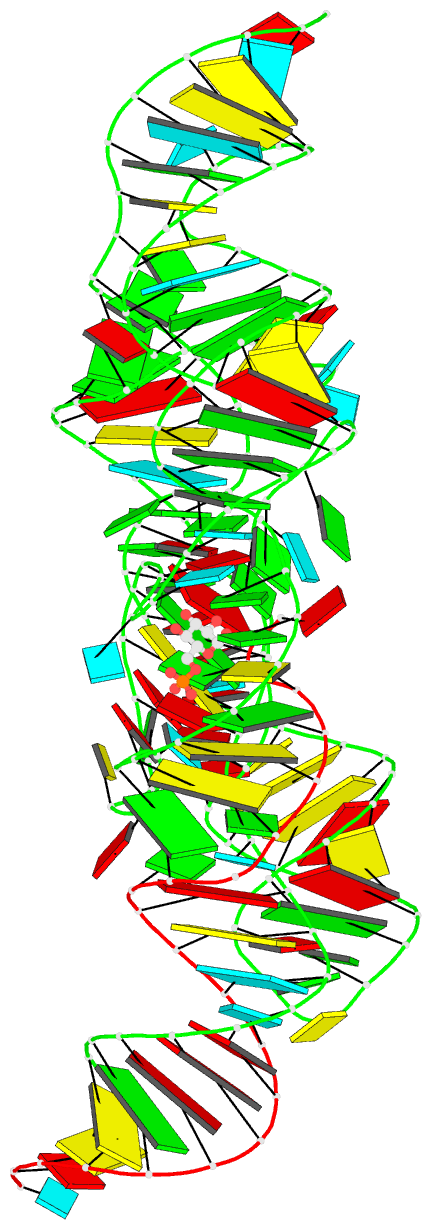
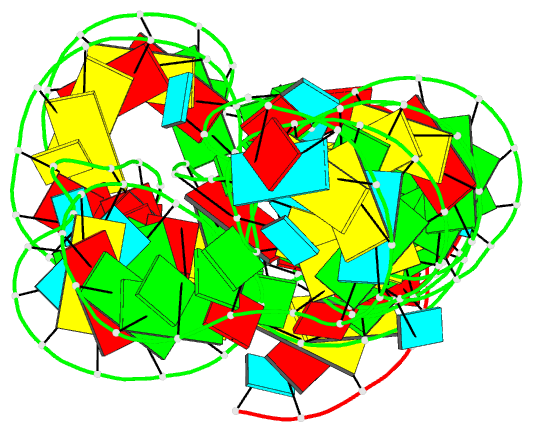
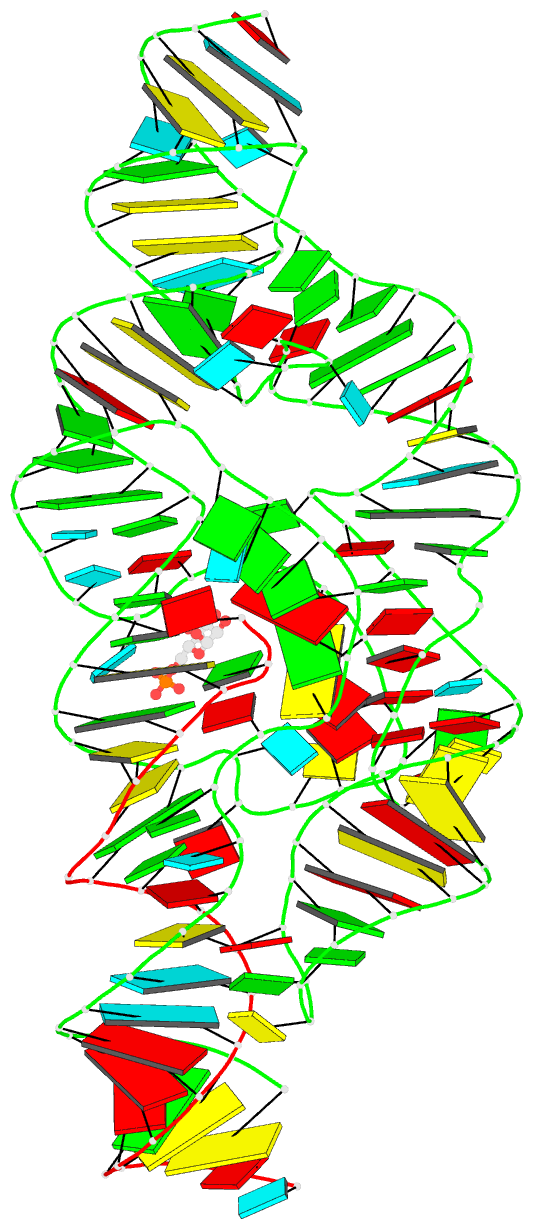
- Summary
- Pre-cleavage state of the thermoanaerobacter tengcongensis glms ribozyme bound to glucose-6-phosphate
- Reference
- Klein DJ, Ferre-D'Amare AR (2006): "Structural basis of glmS ribozyme activation by glucosamine-6-phosphate." Science, 313, 1752-1756. doi: 10.1126/science.1129666.
- Abstract
- The glmS ribozyme is the only natural catalytic RNA known to require a small-molecule activator for catalysis. This catalytic RNA functions as a riboswitch, with activator-dependent RNA cleavage regulating glmS messenger RNA expression. We report crystal structures of the glmS ribozyme in precleavage states that are unliganded or bound to the competitive inhibitor glucose-6-phosphate and in the postcleavage state. All structures superimpose closely, revealing a remarkably rigid RNA that contains a preformed active and coenzyme-binding site. Unlike other riboswitches, the glmS ribozyme binds its activator in an open, solvent-accessible pocket. Our structures suggest that the amine group of the glmS ribozyme-bound coenzyme performs general acid-base and electrostatic catalysis.