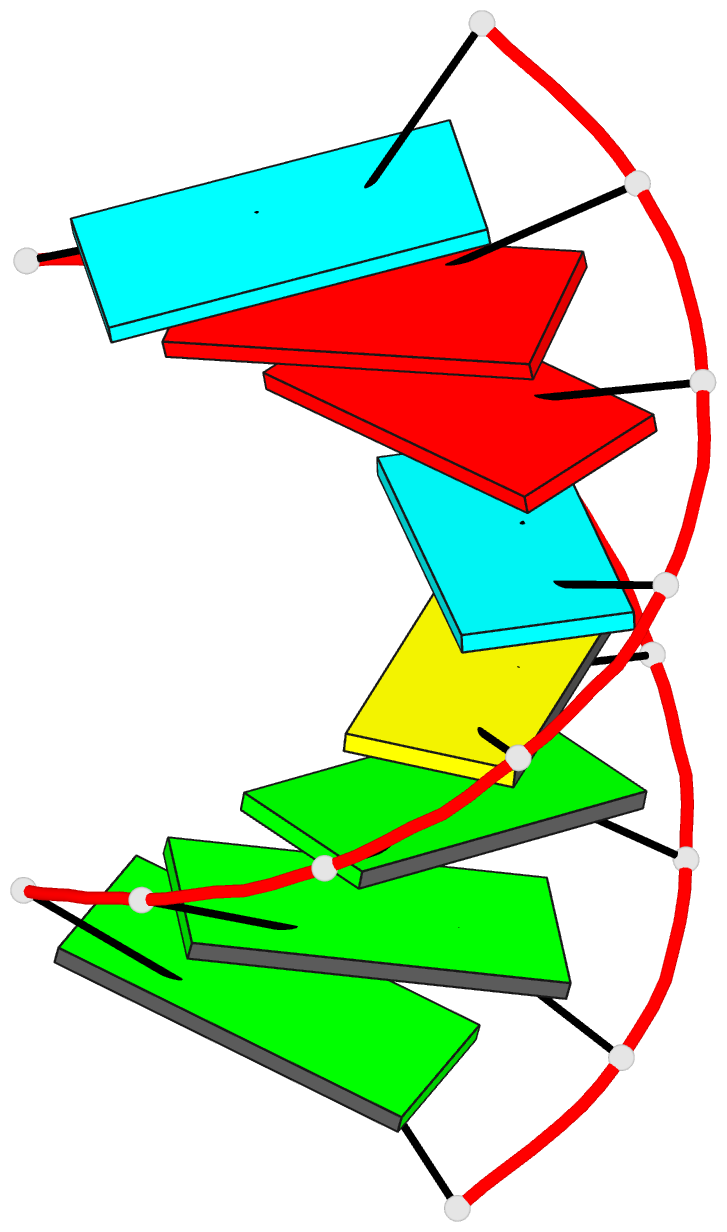
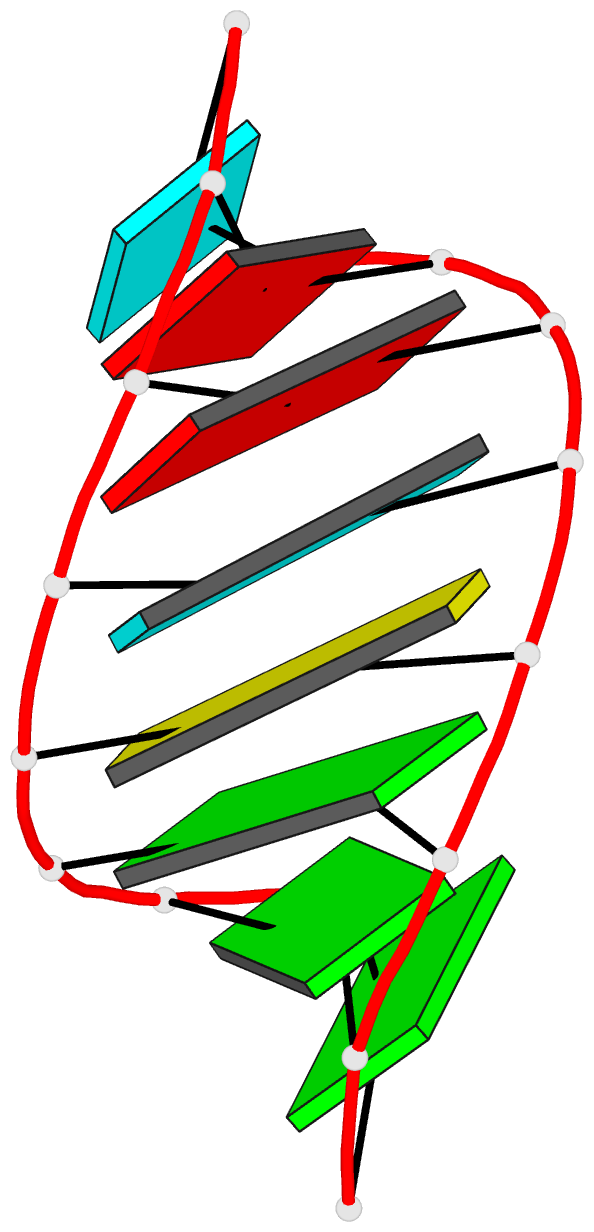
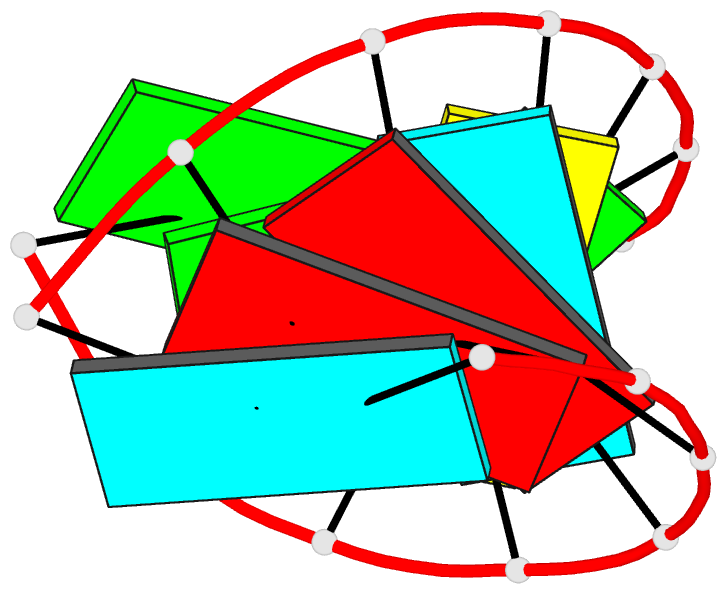
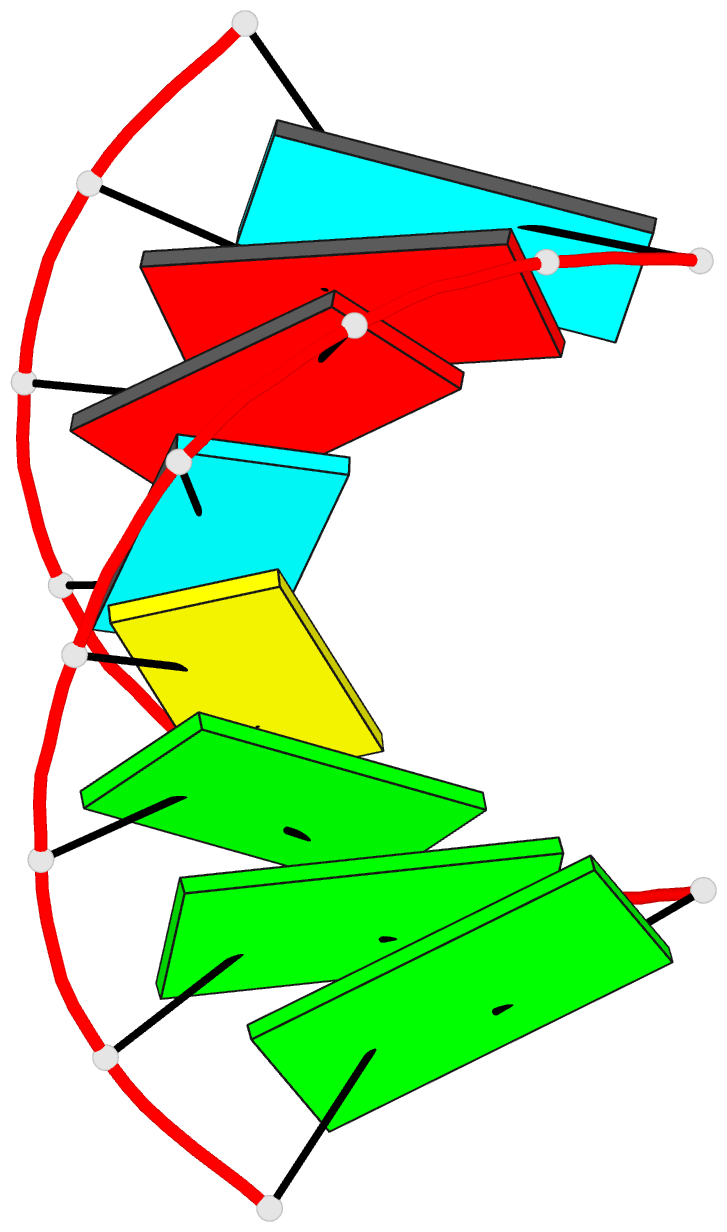
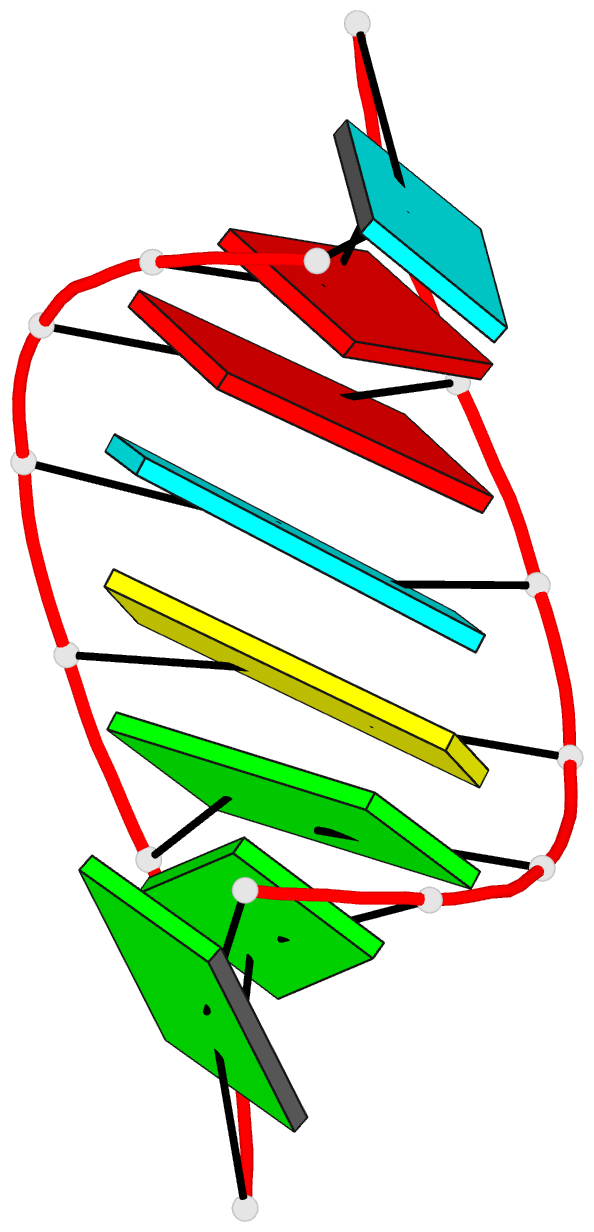
Summary information and primary citation
- PDB-id
- 2gbh; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- NMR structure of stem region of helix-35 of 23s e.coli ribosomal RNA (residues 736-760)
- Reference
- O'Neil-Cabello E, Bryce DL, Nikonowicz EP, Bax A (2004): "Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics." J.Am.Chem.Soc., 126, 66-67. doi: 10.1021/ja038314k.
- Abstract
- A new three-dimensional NMR experiment is described that yields five scalar or dipolar couplings from a single cross-peak between three spins. The method is based on the E.COSY principle and is demonstrated for the H1'-C1'-C2' fragment of ribose sugars in a uniformly 13C-enriched 24-nucleotide RNA stem-loop structure, for which a complete set of couplings was obtained for all nonmodified nucleotides. The values of the isotropic J couplings and the 13C1' and 13C2' chemical shifts define the sugar pucker. Once the sugar pucker is known, the five dipolar couplings between C1'-H1', C2'-H2', H1'-H2', C1'-H2', and C2'-H1', together with C1'-C2', C3'-H3', and C4'-H4' available from standard experiments, can be used to derive the five unknowns that define the local alignment tensor, thereby simultaneously providing information on relative orientation and dynamics of the ribose units. Data indicate rather uniform alignment for all stem nucleotides in the 24-nt stem-loop structure, with only a modest reduction in order for the terminal basepair, but significantly increased mobility in part of the loop region. The method is applicable to proteins, nucleic acids, and carbohydrates, provided 13C enrichment is available.