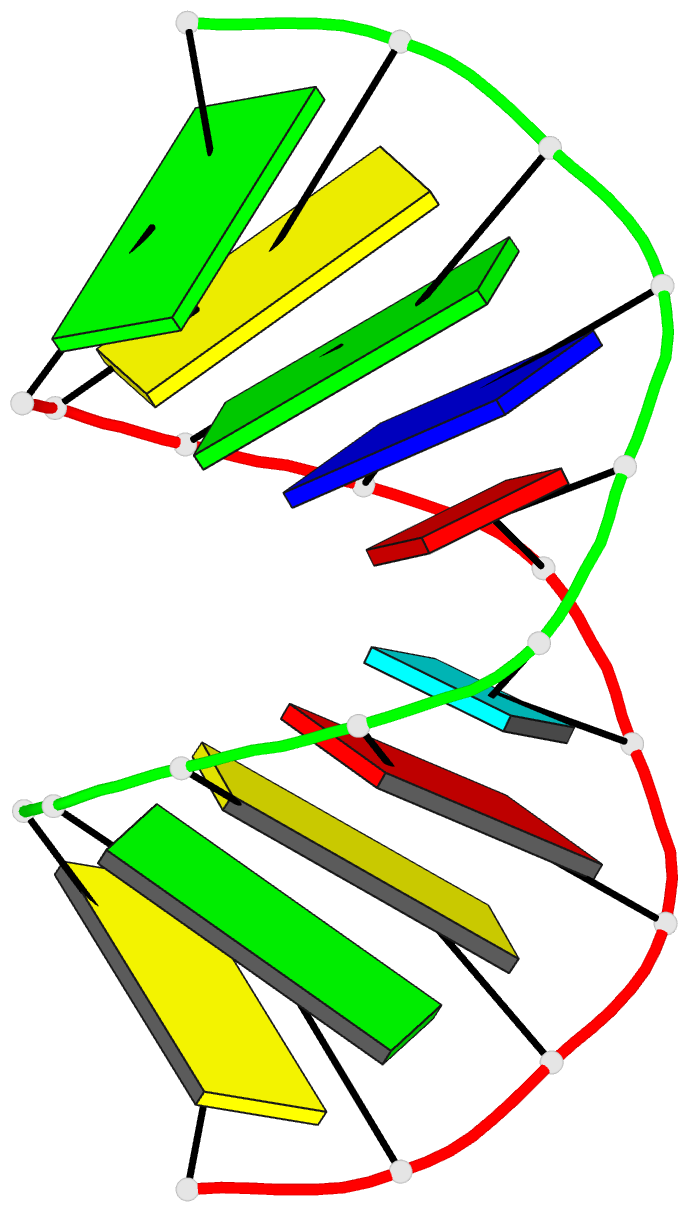
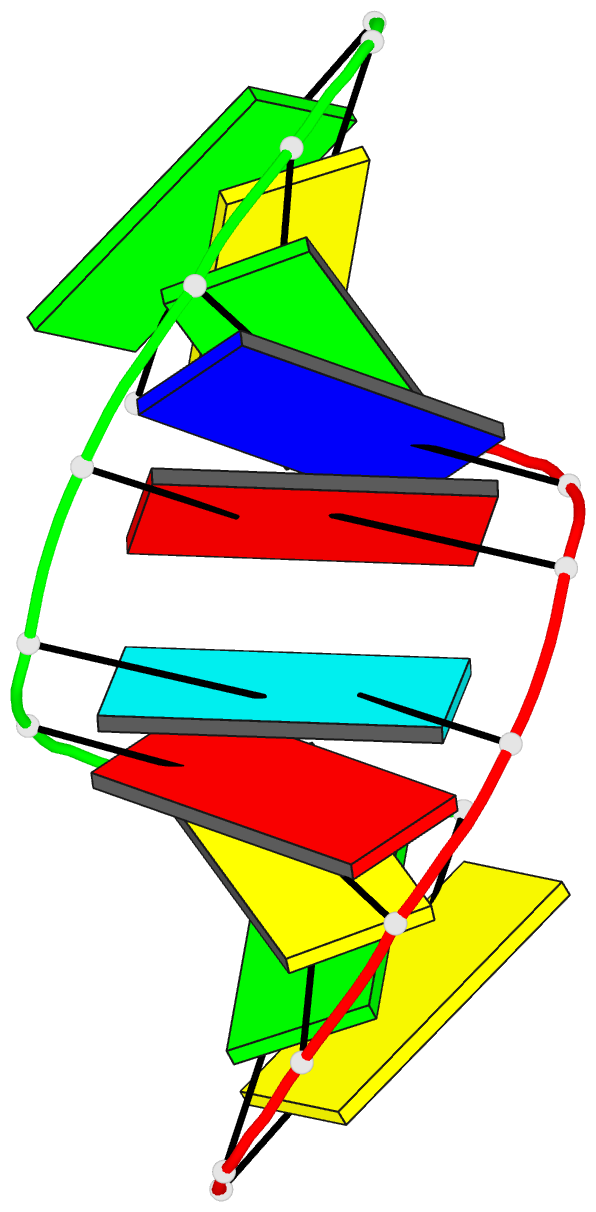
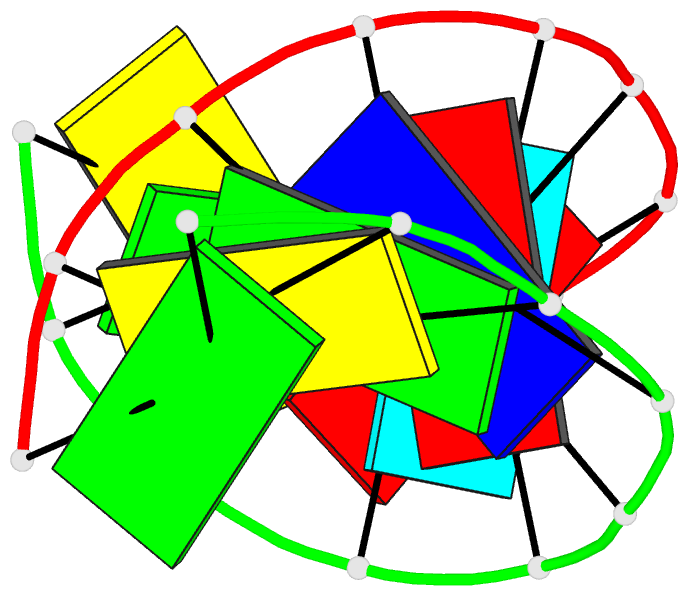
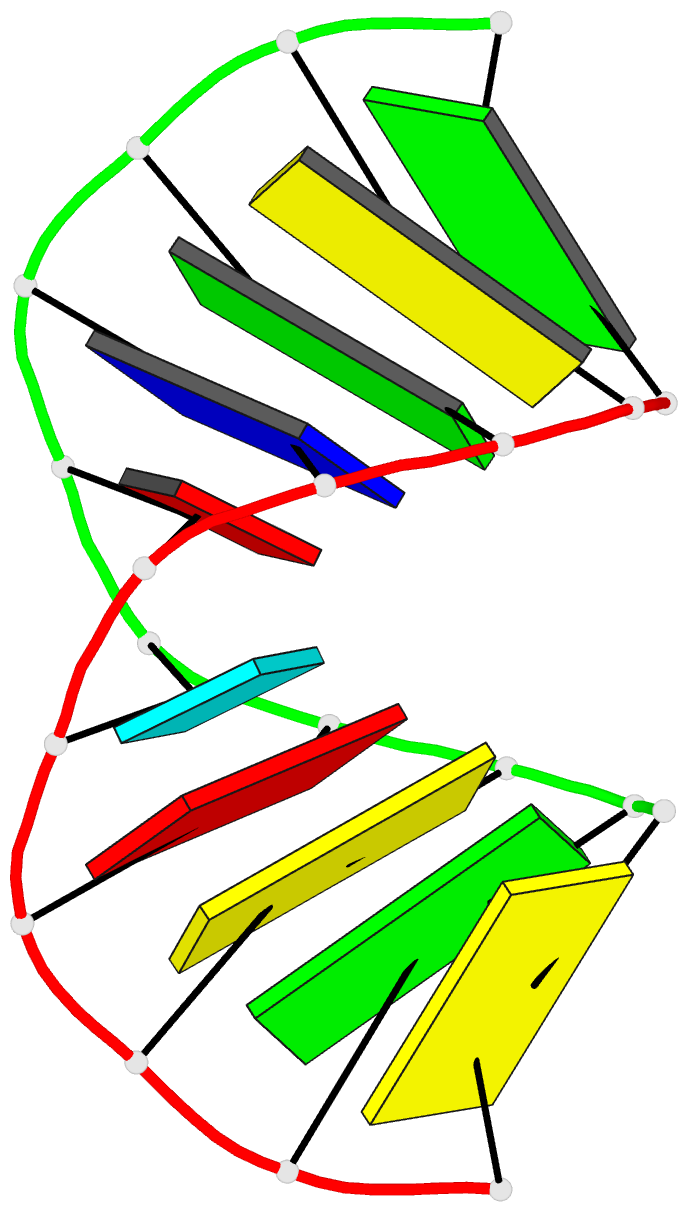
Summary information and primary citation
- PDB-id
- 2fij; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- X-ray (1.19 Å)
- Summary
- Crystal structure analysis of the a-DNA decamer gcgt-2'omea-au-acgc, with incorporated 2'-o-methylated-adenosine (2'omea) and arabino-uridine (au)
- Reference
- Li F, Sarkhel S, Wilds CJ, Wawrzak Z, Prakash TP, Manoharan M, Egli M (2006): "2'-Fluoroarabino- and arabinonucleic acid show different conformations, resulting in deviating RNA affinities and processing of their heteroduplexes with RNA by RNase H." Biochemistry, 45, 4141-4152. doi: 10.1021/bi052322r.
- Abstract
- 2'-Deoxy-2'-fluoro-arabinonucleic acid (FANA) and arabinonucleic acid (ANA) paired to RNA are substrates of RNase H. The conformation of the natural DNA/RNA hybrid substrates appears to be neither A-form nor B-form. Consistent with this, the conformations of FANA and ANA were found to be intermediate between the A- and B-forms. However, FANA opposite RNA is preferred by RNase H over ANA, and the RNA affinity of FANA considerably exceeds that of ANA. By investigating the conformational boundaries of FANA and ANA residues in crystal structures of A- and B-form DNA duplexes at atomic resolution, we demonstrate that FANA and ANA display subtle conformational differences. The structural data provide insight into the structural requirements at the catalytic site of RNase H. They also allow conclusions with regard to the relative importance of stereoelectronic effects and hydration as modulators of RNA affinity.