Summary information and primary citation
- PDB-id
- 2awe; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (2.1 Å)
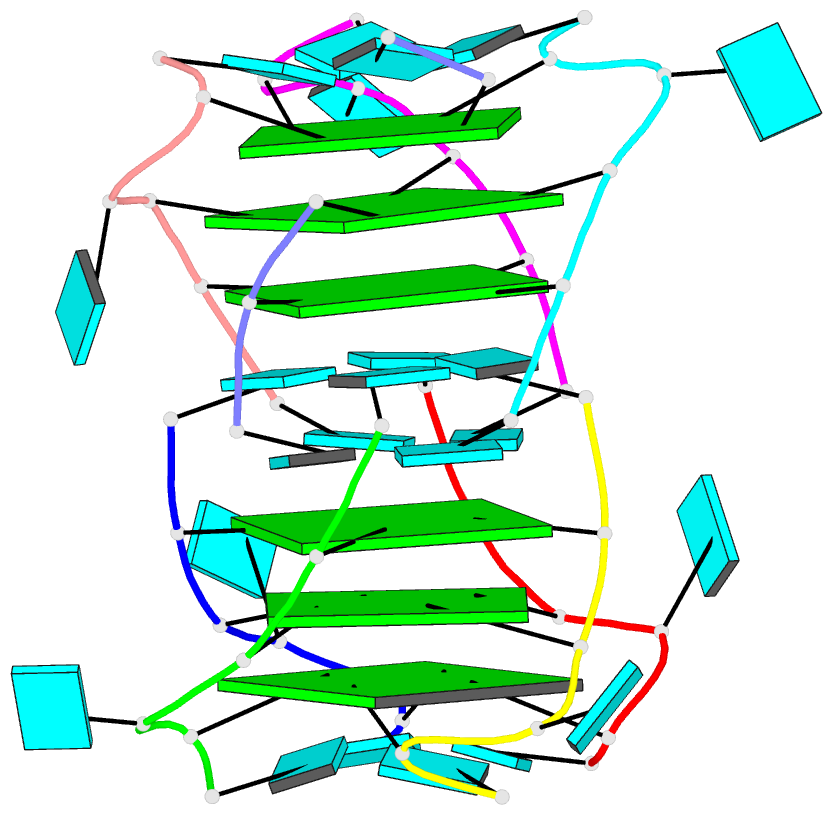
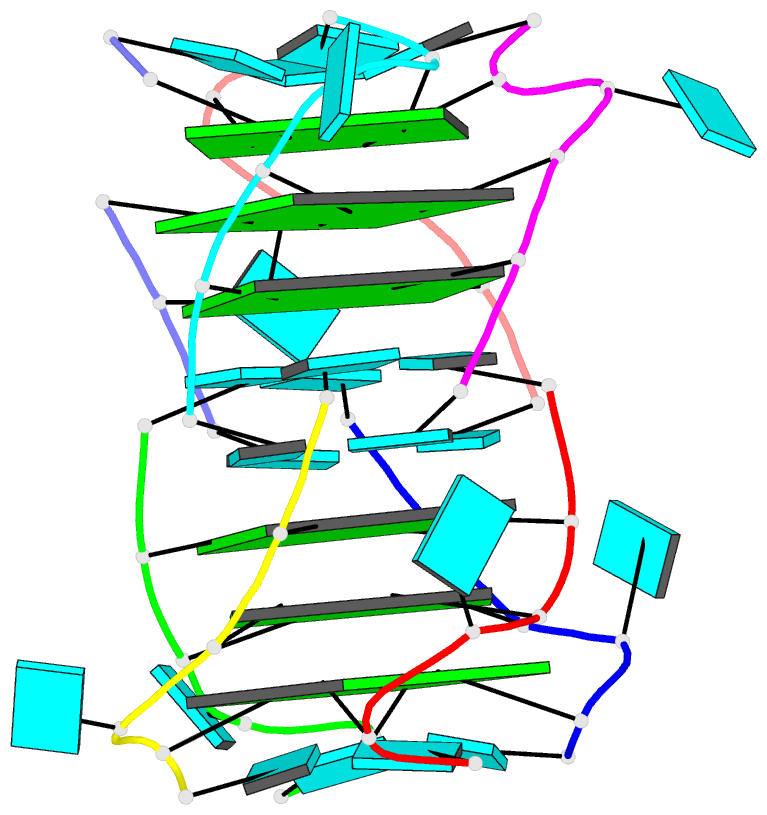
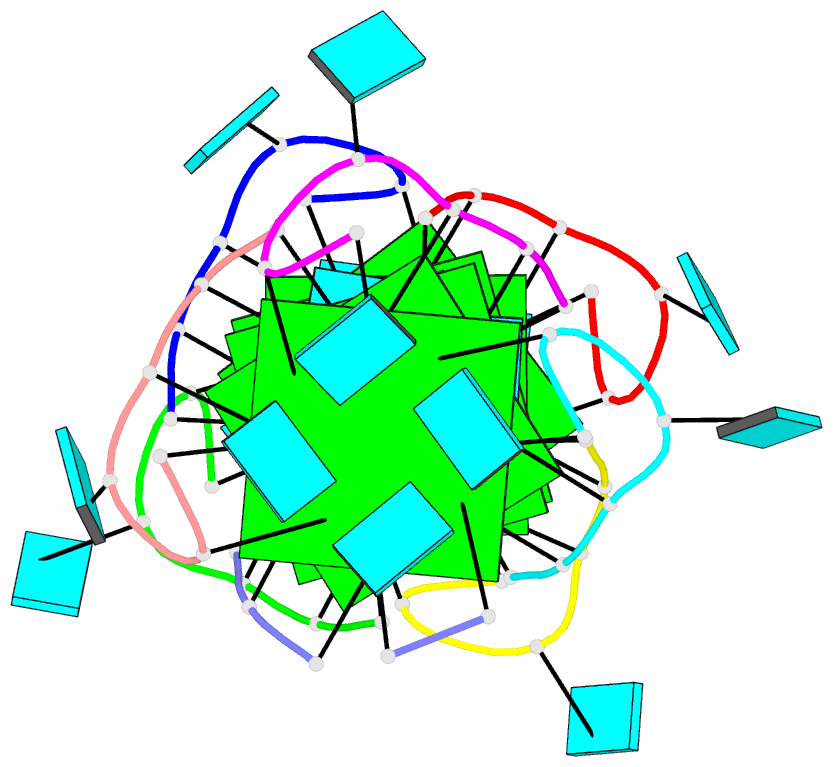
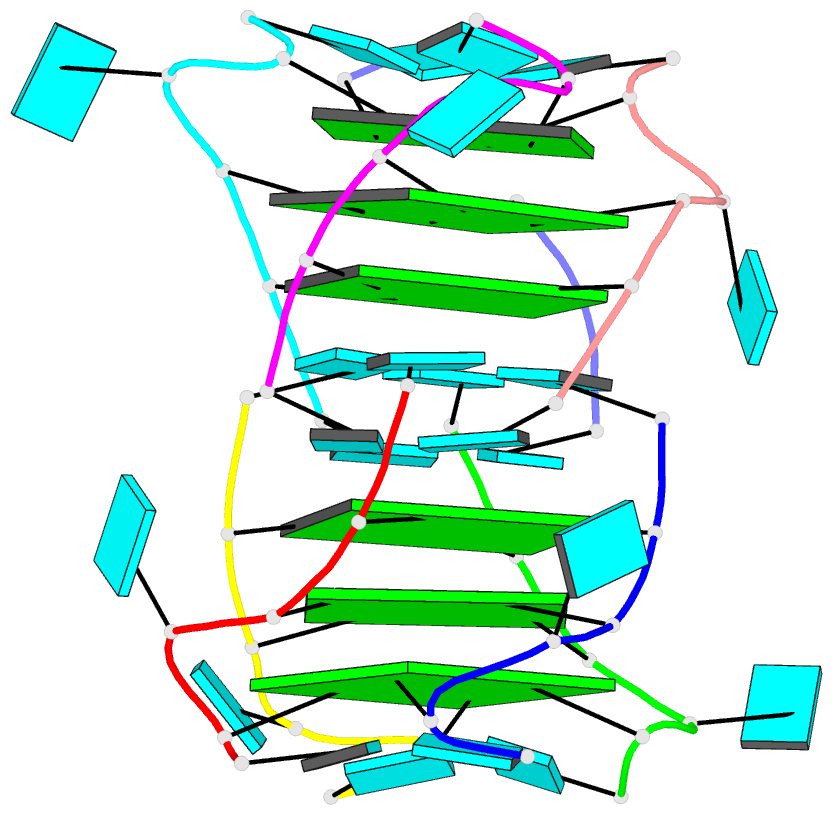
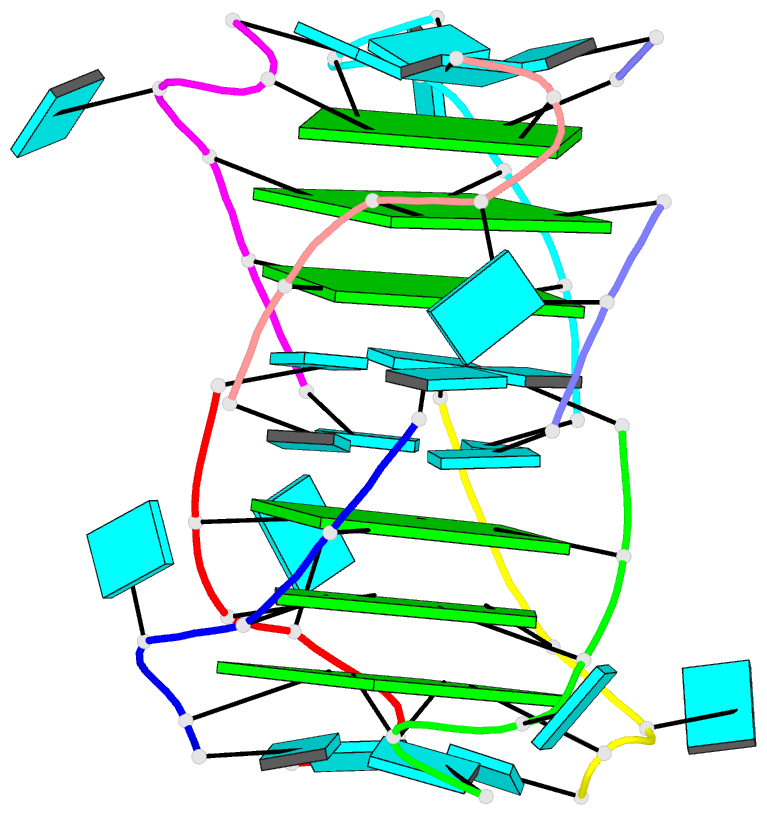
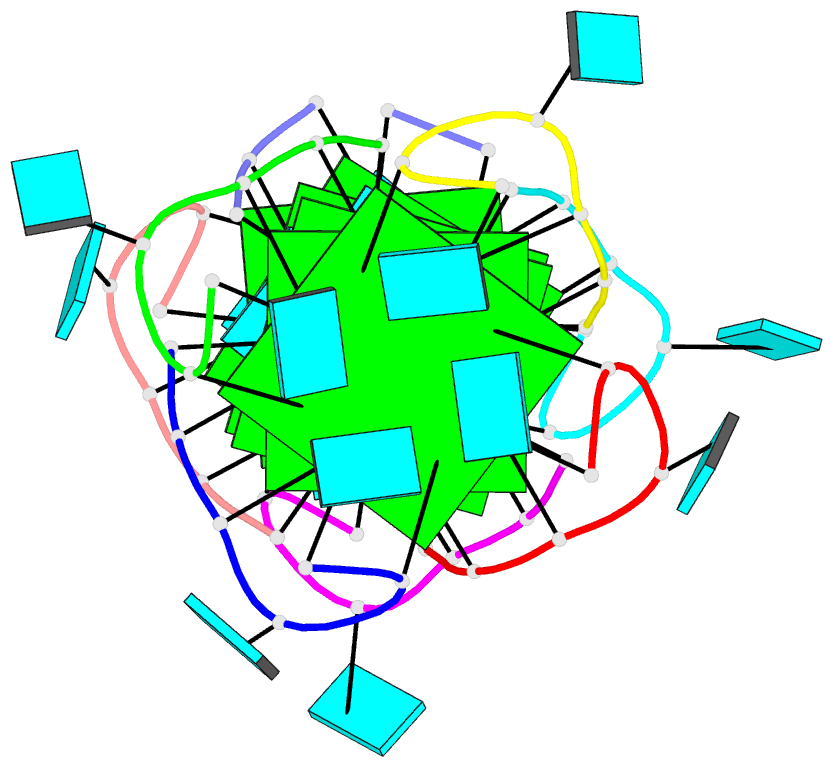
- Summary
- Base-tetrad swapping results in dimerization of RNA quadruplexes: implications for formation of i-motif RNA octaplex
- Reference
- Pan B, Shi K, Sundaralingam M (2006): "Base-tetrad swapping results in dimerization of RNA quadruplexes: implications for formation of the i-motif RNA octaplex." Proc.Natl.Acad.Sci.Usa, 103, 3130-3134. doi: 10.1073/pnas.0507730103.
- Abstract
- Nucleic acids adopt different multistranded helical architectures to perform various biological functions. Here, we report a crystal structure of an RNA quadruplex containing "base-tetrad swapping" and bulged nucleotide at 2.1-Angstroms resolution. The base-tetrad swapping results in a dimer of quadruplexes with an intercalated octaplex fragment at the 5' end junction. The intercalated base tetrads provide the basic repeat unit for constructing a model of intercalated RNA octaplex. The model we obtained shows fundamentally different characteristics from duplex, triplex, and quadruplex. We also observed two different orientations of bulged uridine residues that are related to the interaction with surroundings. This structural evidence reflects the conformational flexibility of bulged nucleotides in RNA quadruplexes and implies the potential roles of bulged nucleotides as recognition and interaction sites in RNA-protein and RNA-RNA interactions.