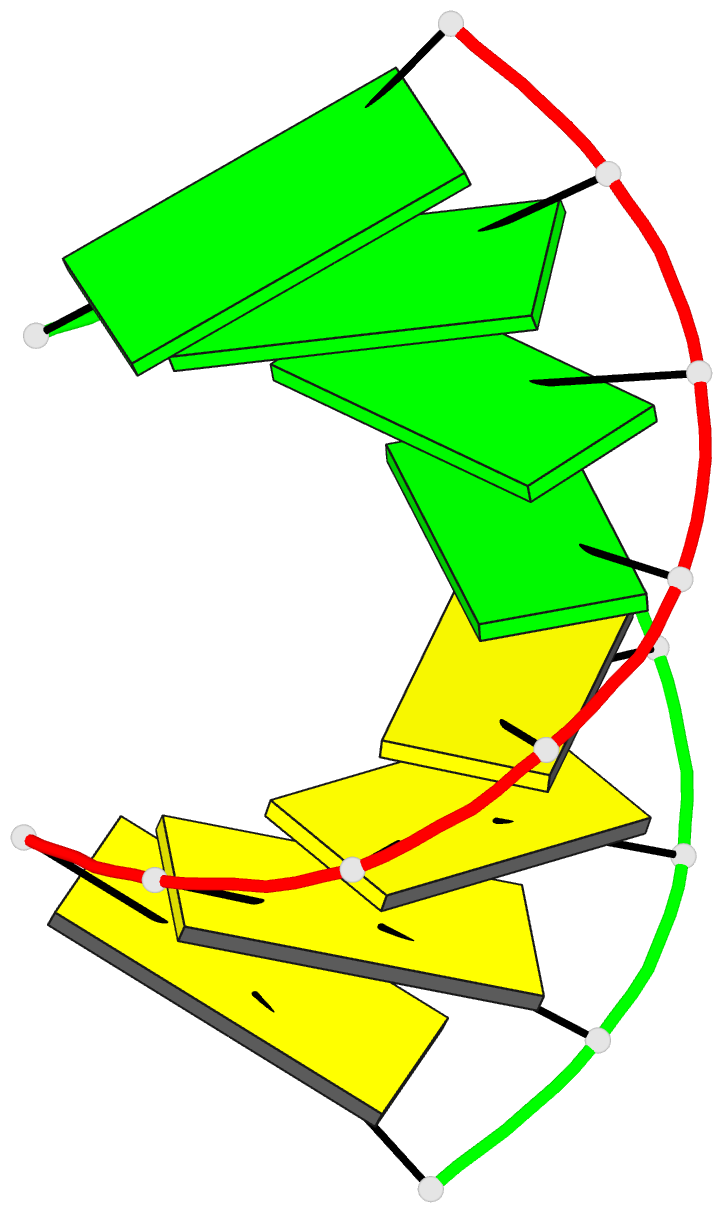
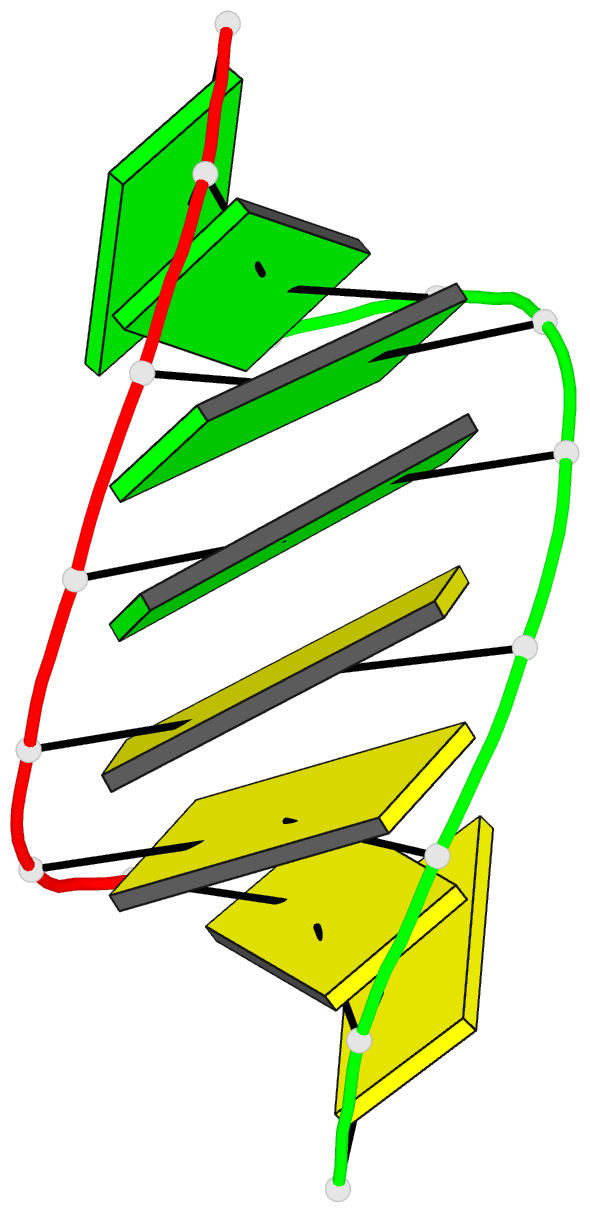
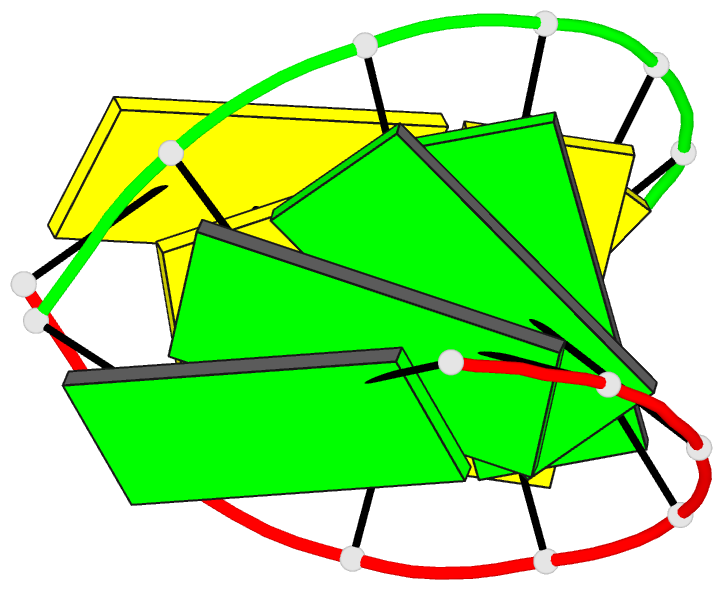
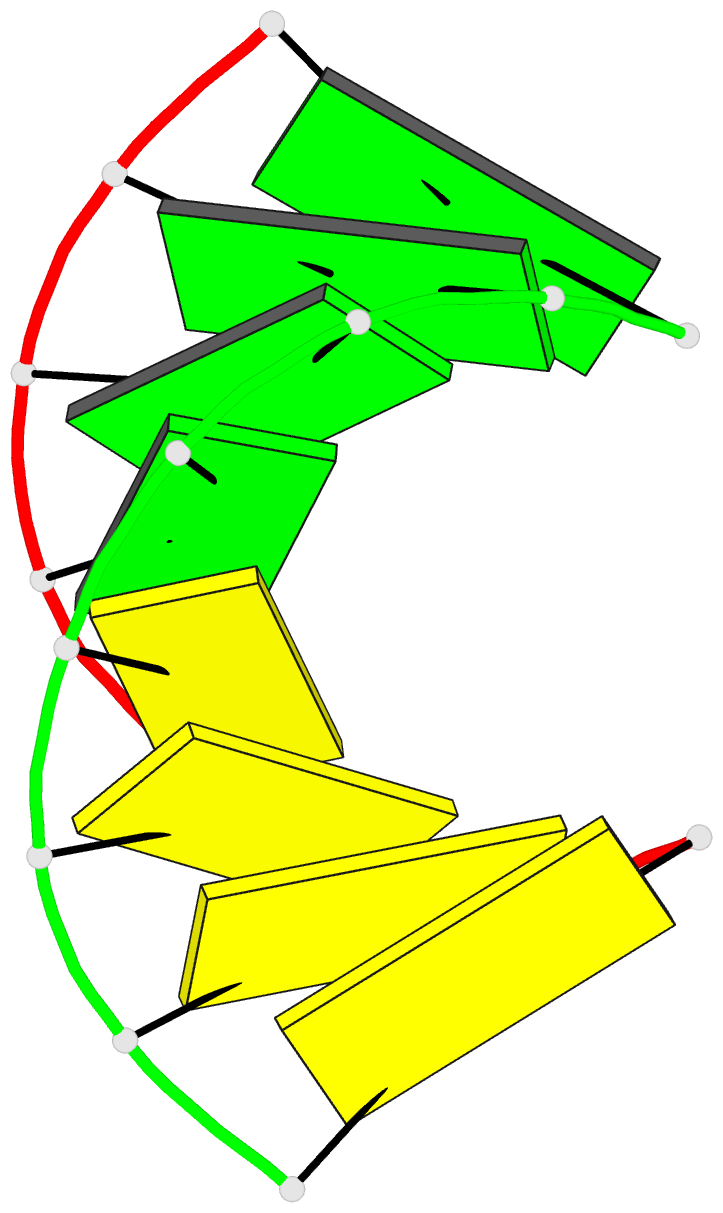
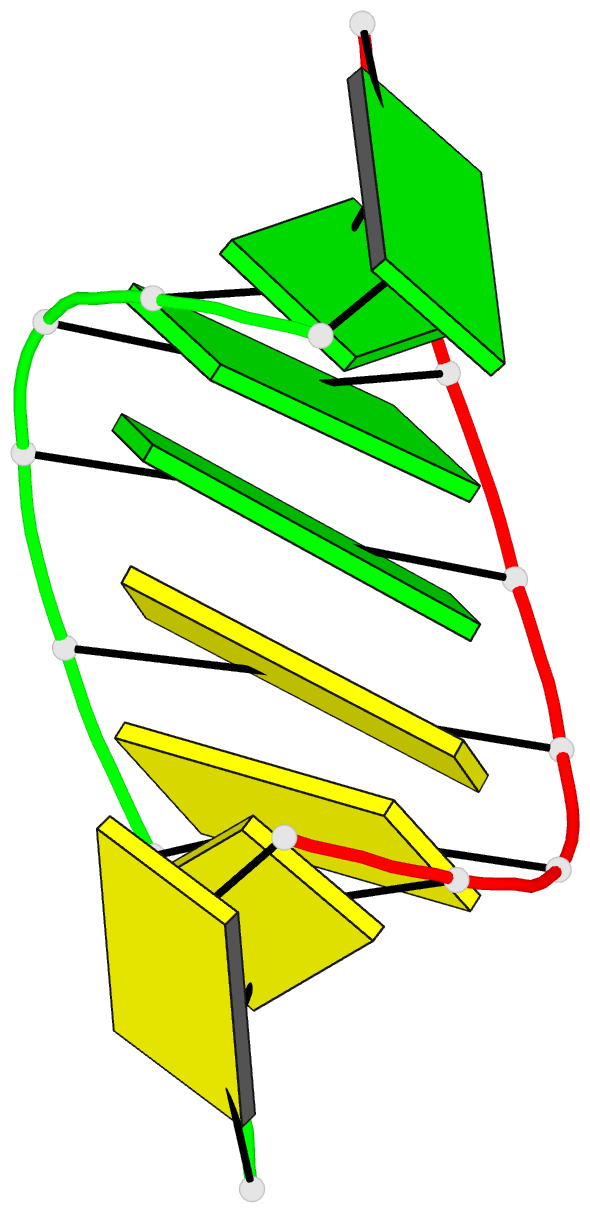
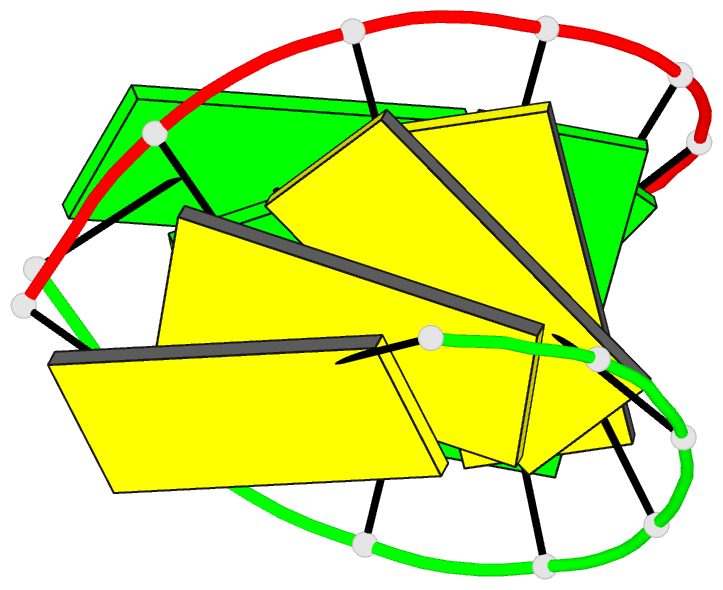
Summary information and primary citation
- PDB-id
- 2a0p; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (1.95 Å)
- Summary
- Crystal structure of RNA oligomer containing 4'-thioribose
- Reference
- Haeberli P, Berger I, Pallan PS, Egli M (2005): "Syntheses of 4'-thioribonucleosides and thermodynamic stability and crystal structure of RNA oligomers with incorporated 4'-thiocytosine." Nucleic Acids Res., 33, 3965-3975. doi: 10.1093/nar/gki704.
- Abstract
- A facile synthetic route for the 4'-thioribonucleoside building block (4'S)N (N = U, C, A and G) with the ribose O4' replaced by sulfur is presented. Conversion of l-lyxose to 1,5-di-O-acetyl-2,3-di-O-benzoyl-4-thio-d-ribofuranose was achieved via an efficient four-step synthesis with high yield. Conversion of the thiosugar into the four ribonucleoside phosphoramidite building blocks was accomplished with additional four steps in each case. Incorporation of 4'-thiocytidines into oligoribonucleotides improved the thermal stability of the corresponding duplexes by approximately 1 degrees C per modification, irrespective of whether the strand contained a single modification or a consecutive stretch of (4'S)C residues. The gain in thermodynamic stability is comparable to that observed with oligoribonucleotides containing 2'-O-methylated residues. To establish potential conformational changes in RNA as a result of the 4'-thio modification and to better understand the origins of the observed stability changes, the crystal structure of the oligonucleotide 5'-r(CC(4'S)CCGGGG) was determined and analyzed using the previously solved structure of the native RNA octamer as a reference. The two 4'-thioriboses adopt conformations that are very similar to the C3'-endo pucker observed for the corresponding sugars in the native duplex. Subtle changes in the local geometry of the modified duplex are mostly due to the larger radius of sulfur compared to oxygen or appear to be lattice-induced. The significantly increased RNA affinity of 4'-thio-modified RNA relative to RNA, and the relatively minor conformational changes caused by the modification render this nucleic acid analog an interesting candidate for in vitro and in vivo applications, including use in RNA interference (RNAi), antisense, ribozyme, decoy and aptamer technologies.