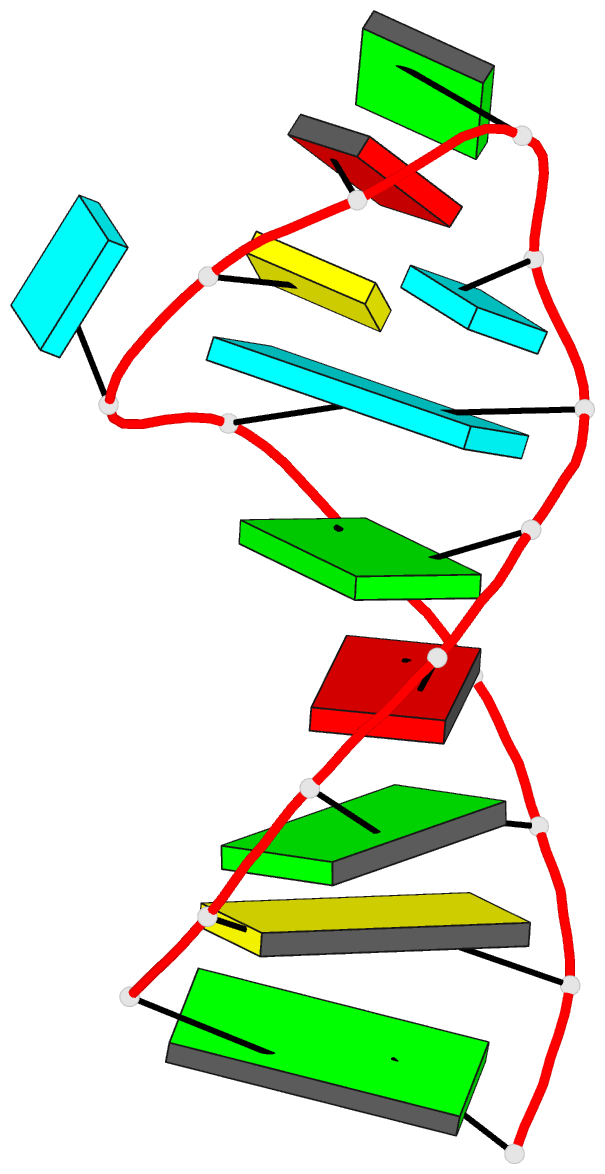
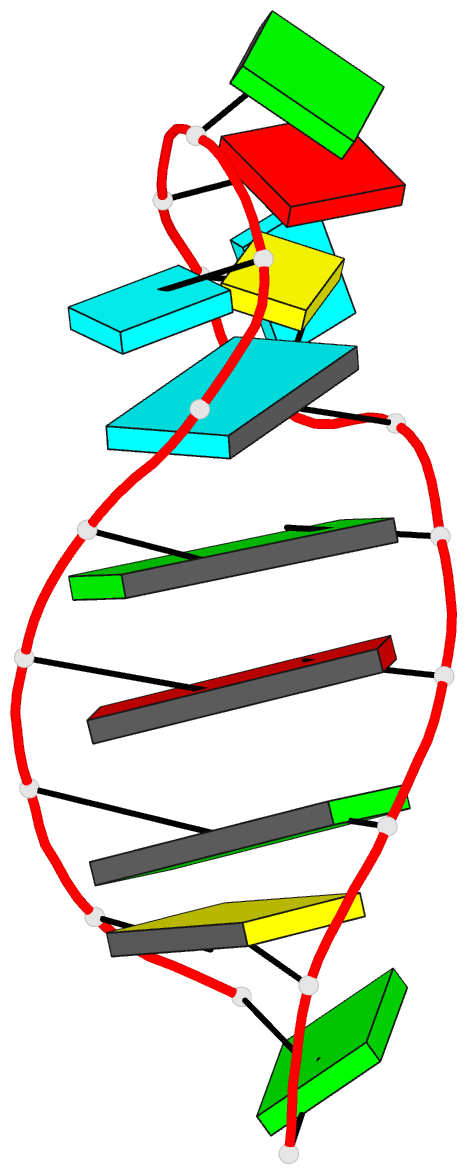
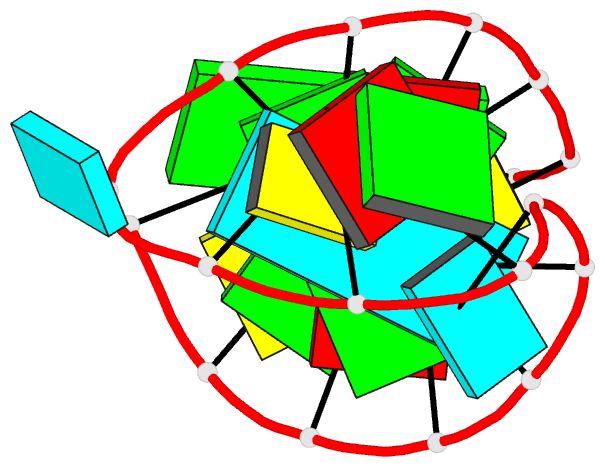
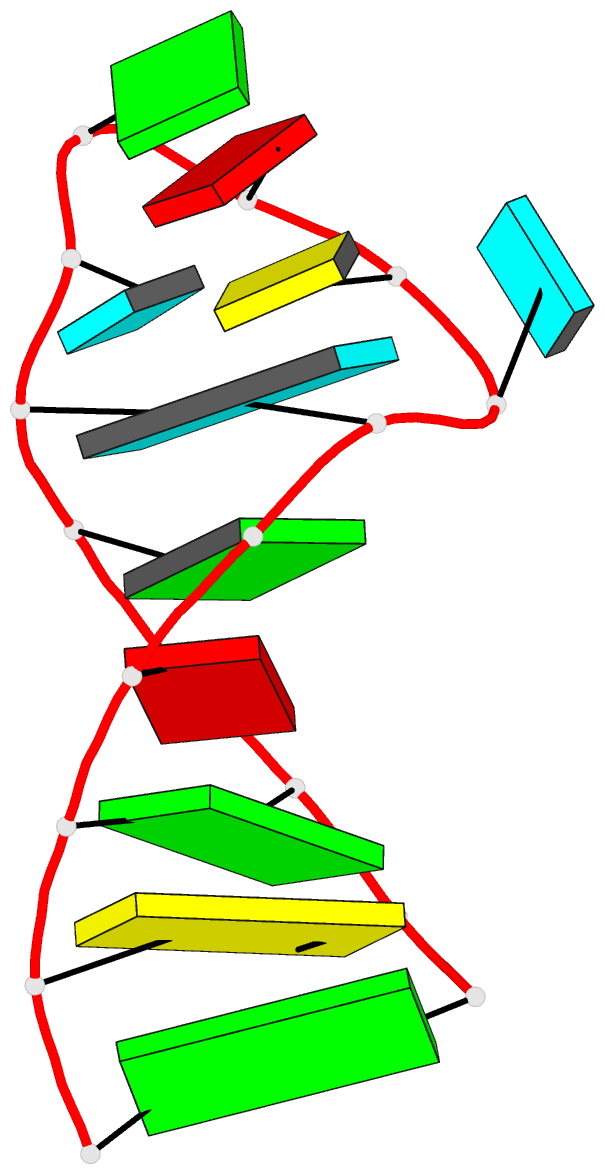
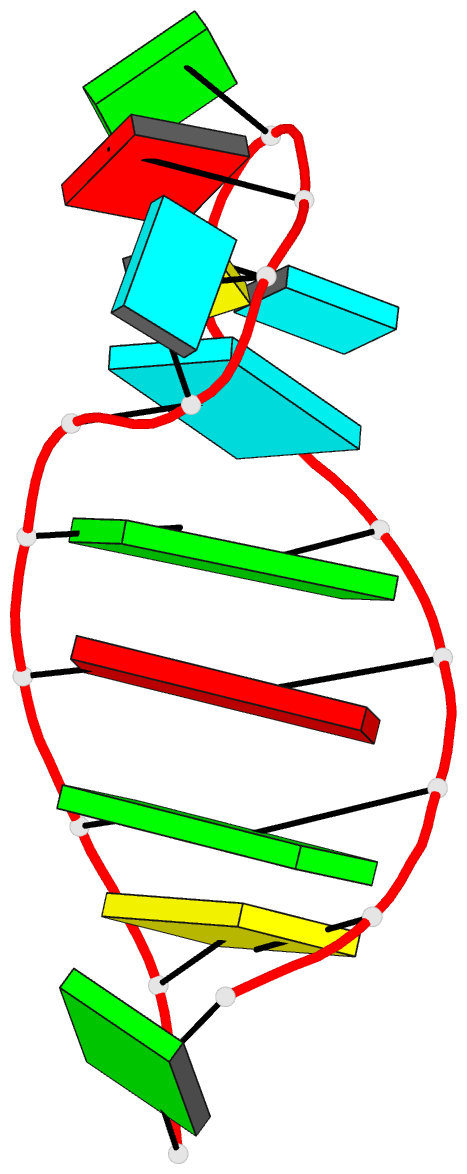
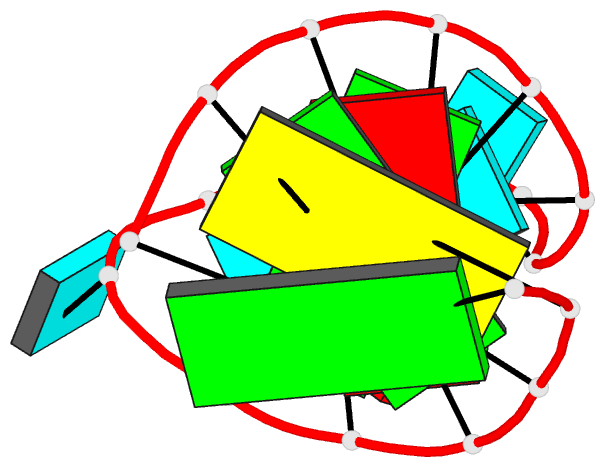
Summary information and primary citation
- PDB-id
-
1yn1;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Solution structure of the vs ribozyme stem-loop v in
the presence of mgcl2
- Reference
-
Campbell DO, Bouchard P, Desjardins G, Legault P (2006):
"NMR
structure of varkud satellite ribozyme stem-loop v in the
presence of magnesium ions and localization of
metal-binding sites." Biochemistry,
45, 10591-10605. doi: 10.1021/bi0607150.
- Abstract
- In the Neurospora VS ribozyme, magnesium ions
facilitate formation of a loop-loop interaction between
stem-loops I and V, which is important for recognition and
activation of the stem-loop I substrate. Here, we present
the high-resolution NMR structure of stem-loop V (SL5) in
the presence of Mg(2+) (SL5(Mg)) and demonstrate that
Mg(2+) induces a conformational change in which the SL5
loop adopts a compact structure with most characteristics
of canonical U-turn structures. Divalent cation-binding
sites were probed with Mn(2+)-induced paramagnetic line
broadening and intermolecular NOEs to Co(NH(3))(6)(3+).
Structural modeling of Mn(H(2)O)(6)(2+) in SL5(Mg) revealed
four divalent cation-binding sites in the loop. Sites 1, 3,
and 4 are located in the major groove near multiple
phosphate groups, whereas site 2 is adjacent to N7 of G697
and N7 of A698 in the minor groove. Cation-binding sites
equivalent to sites 1-3 in SL5 are present in other U-turn
motifs, and these metal-binding sites may represent a
common feature of the U-turn fold. Although magnesium ions
affect the loop conformation, they do not significantly
change the conformation of residues 697-699 involved in the
proposed Watson-Crick base pairs with stem-loop I. In both
the presence and the absence of Mg(2+), G697, A698, and
C699 adopt an A-form structure that exposes their
Watson-Crick faces, and this is compatible with their
proposed interaction with stem-loop I. In SL5(Mg), however,
U700 becomes exposed on the minor groove face of the loop
in the proximity of the bases of G697, A698, and C699,
suggesting that the Mg(2+)-bound conformation of stem-loop
V allows additional contacts with stem-loop I. These
studies improve our understanding of the role of Mg(2+) in
U-turn structures and in substrate recognition by the VS
ribozyme.