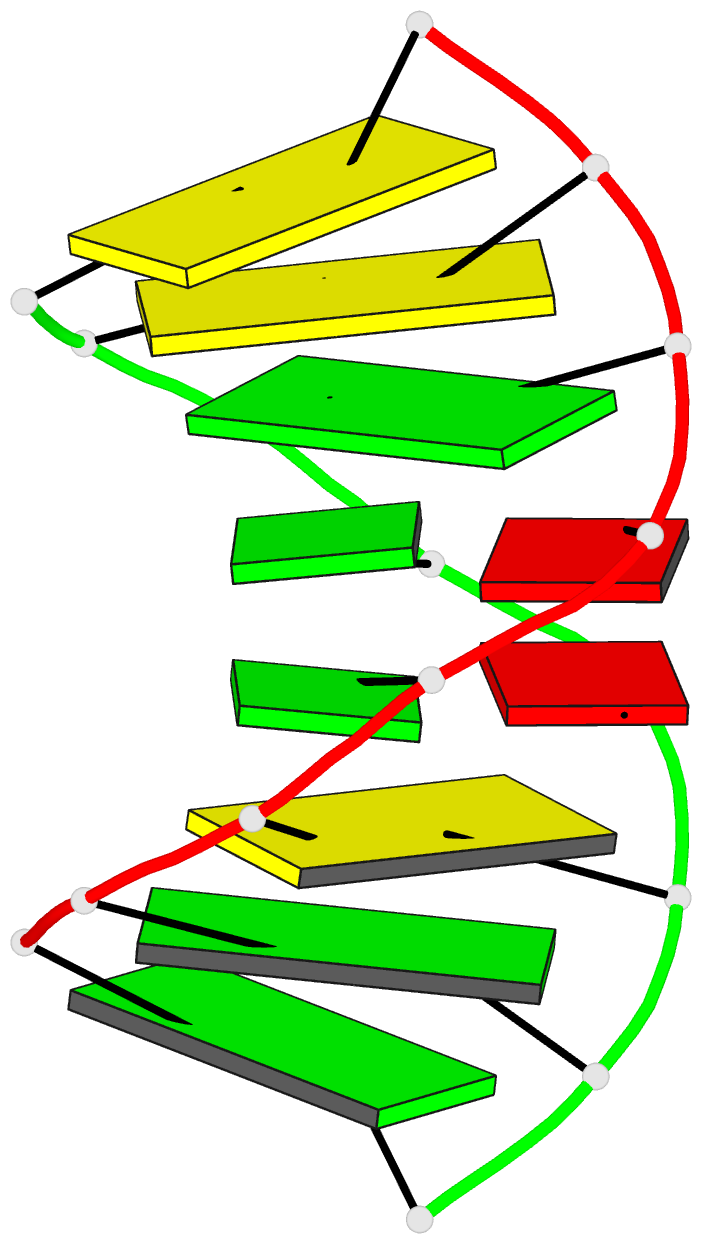
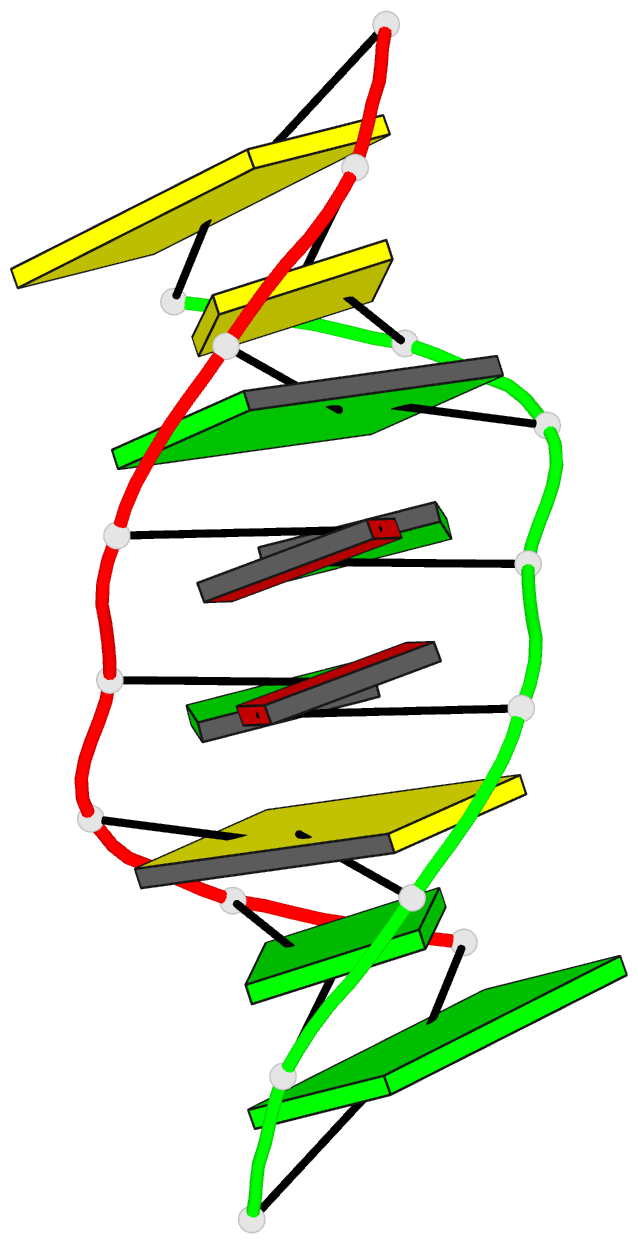
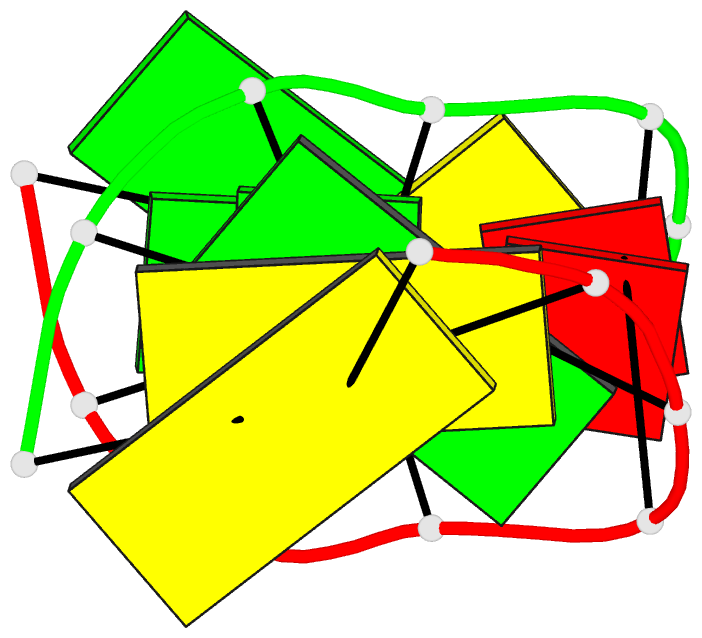
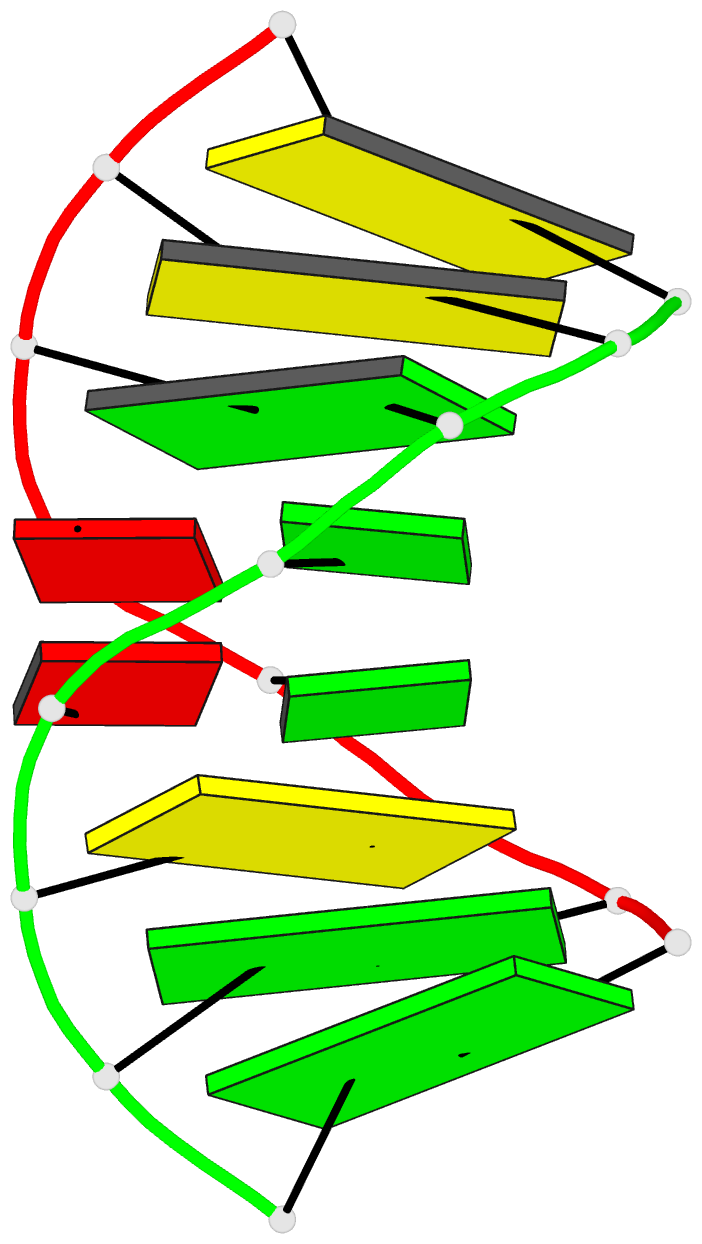
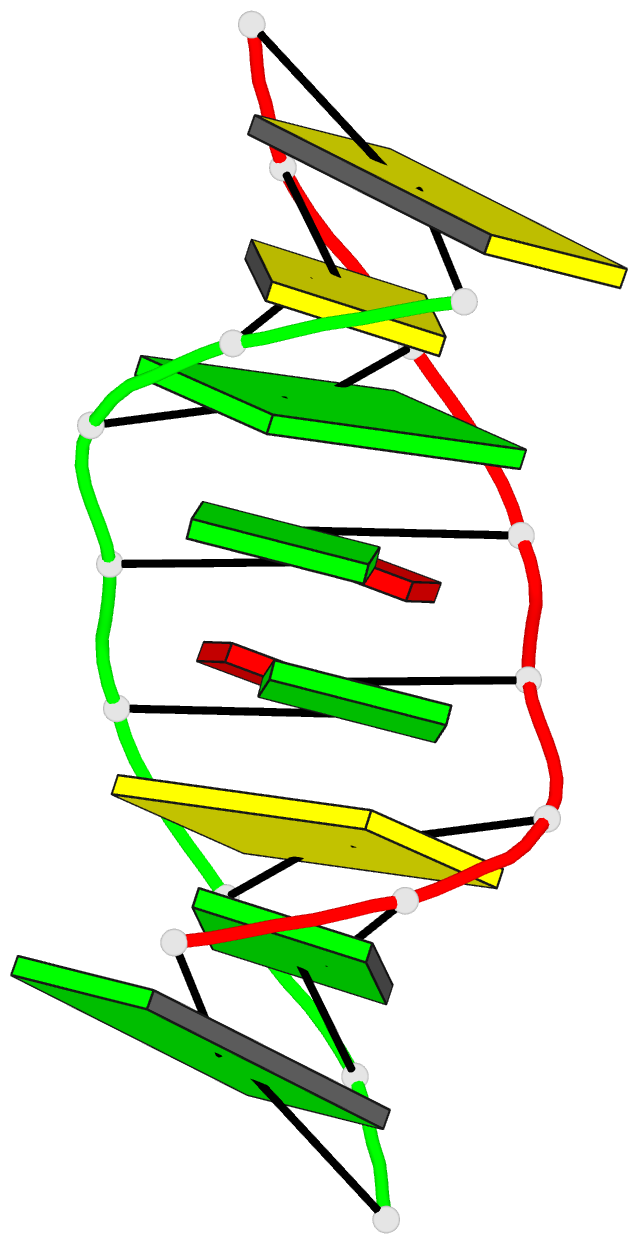
Summary information and primary citation
- PDB-id
- 1yfv; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Structure of (5'-r(gp*gp*cp*gp*ap*gp*cp*c)-3')2 by 2-d NMR, 1 structure
- Reference
- SantaLucia Jr J, Turner DH (1993): "Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics." Biochemistry, 32, 12612-12623. doi: 10.1021/bi00210a009.
- Abstract
- The duplex (rGGCGAGCC)2 contains tandem G x A mismatches--a common motif in the secondary structures of biological RNAs. The three-dimensional structure of (rGGCGAGCC)2 was derived using molecular dynamics and energy minimization with NMR-derived restraints for 78 interproton distances (per strand), 18 hydrogen bonds for the six Watson-Crick G x C pairs, and 26 dihedral angles (per strand). The G x A mismatch structures are similar to those observed in a DNA duplex [Li, Y., Zon, G., & Wilson, W. D. (1991) Proc. Natl. Acad. Sci. U.S.A. 80, 26-30] and an RNA hairpin [Heus, H. A., & Pardi, A. (1991) Science 253, 191-193], with hydrogen bonds from guanine 2-amino and N3 to adenine N7 and 6-amino, respectively. The other G 2-amino and A 6-amino protons are within hydrogen-bonding distance of a phosphate oxygen and 2'-oxygen, respectively. Strong interstrand A-A and G-G stacking is observed between the G x A mismatches. This contrasts with the poor stacking observed between the G x A mismatches and closing G x C base pairs. The stems are basically A-form with all bases in the anti conformation and all nonterminal sugars in the C3'-endo conformation. The structure rationalizes previous thermodynamic, circular dichroism, and imino proton NMR results and suggests tandem G x A mismatches in RNA may provide a contact site for tertiary interactions.