Summary information and primary citation
- PDB-id
- 1qbp; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- X-ray (2.1 Å)
- Summary
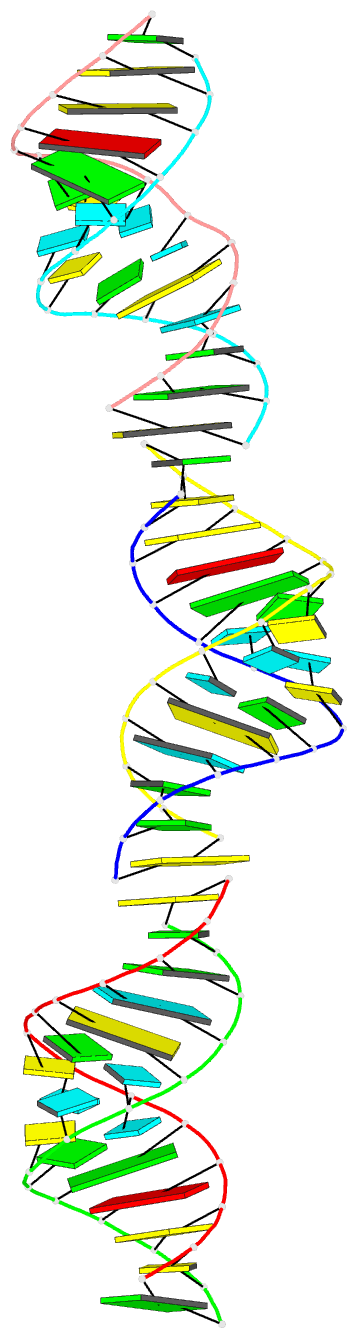
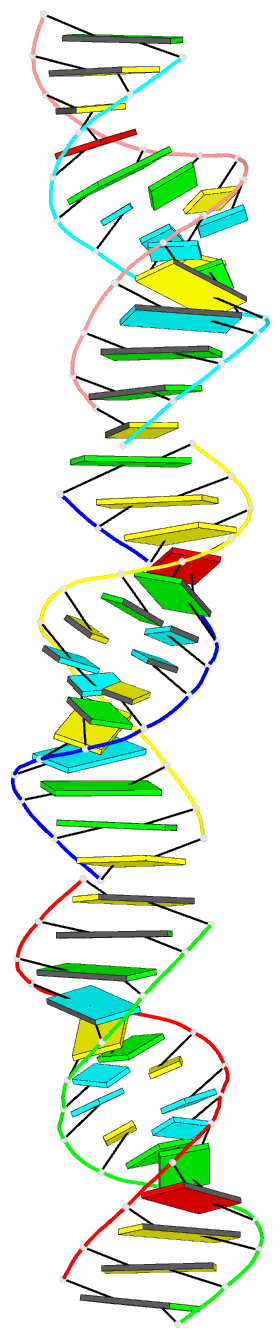
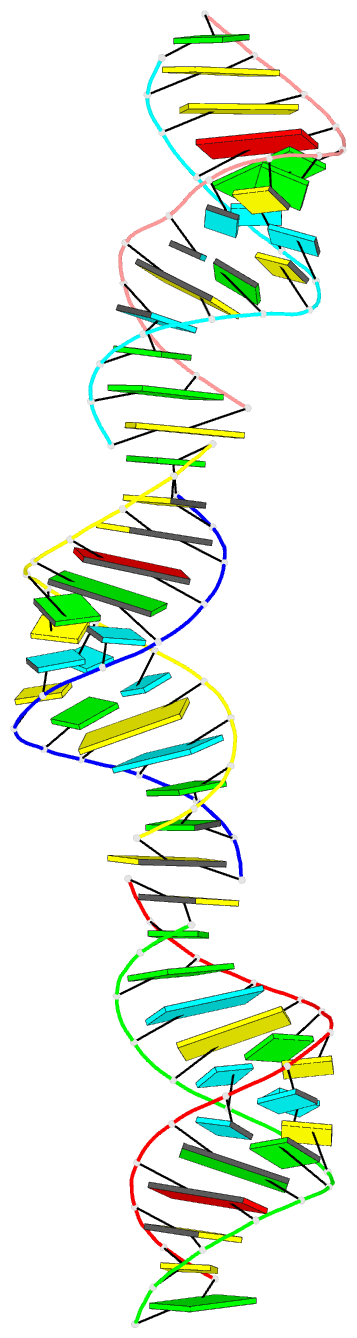
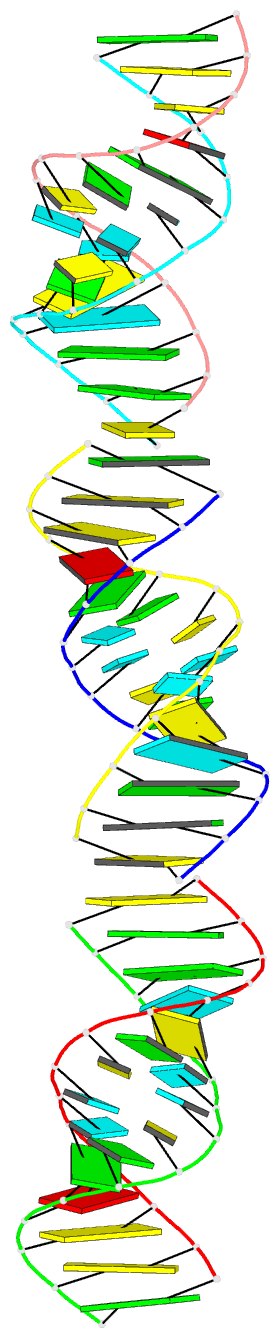
- Crystal structure of a brominated RNA helix with four mismatched base pairs
- Reference
- Anderson AC, O'Neil RH, Filman DJ, Frederick CA (1999): "Crystal structure of a brominated RNA helix with four mismatched base pairs: An investigation into RNA conformational variability." Biochemistry, 38, 12577-12585. doi: 10.1021/bi9904508.
- Abstract
- The X-ray crystal structure of a brominated RNA helix with four mismatched base pairs and sequence r(UG(Br)C(Br)CAGUUCGCUGGC)(2) was determined to 2.1 A using the methods of multiwavelength anomalous diffraction (MAD) applied to the bromine K-absorption edge. There are three molecules in the asymmetric unit with unique crystal-packing environments, revealing true conformational variability at high resolution for this sequence. The structure shows that the sequence itself does not define a consistent pattern of solvent molecules, with the exception of the mismatched base pairs, implying that specific RNA-protein interactions would occur only with the nucleotides. There are a number of significant tertiary interactions, some of which are a result of the brominated base pairs and others that are directly mediated by the RNA 2' hydroxyl groups. The mismatched base pairs exhibit a solvent network as well as a stacking pattern with their nearest neighbors that validate previous thermodynamic analysis.