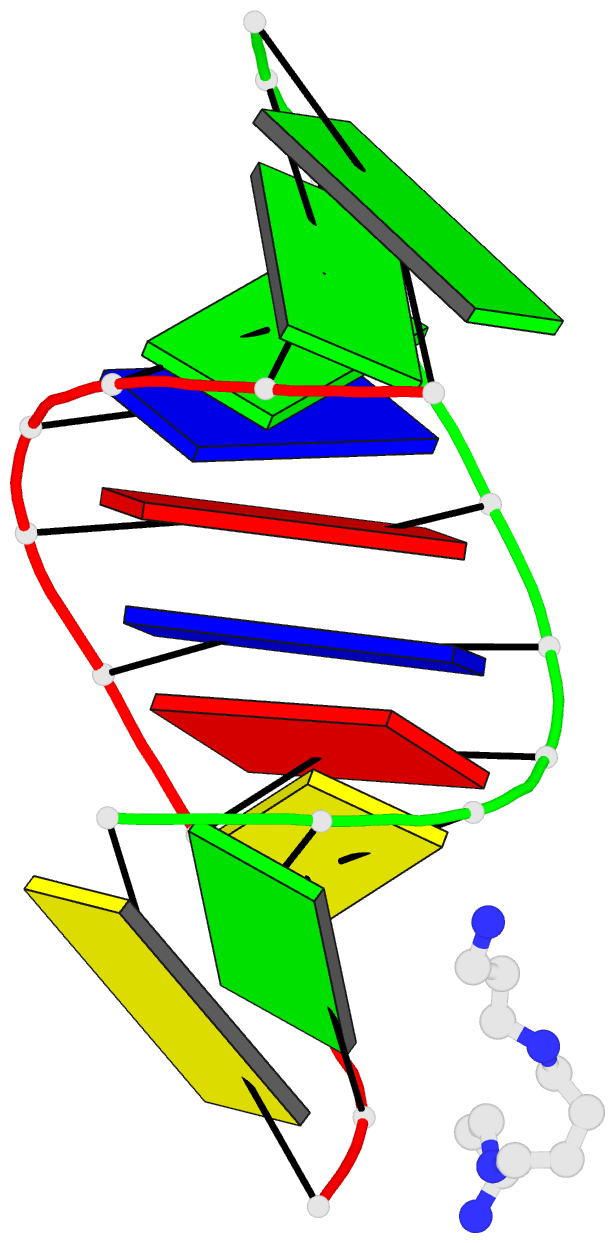
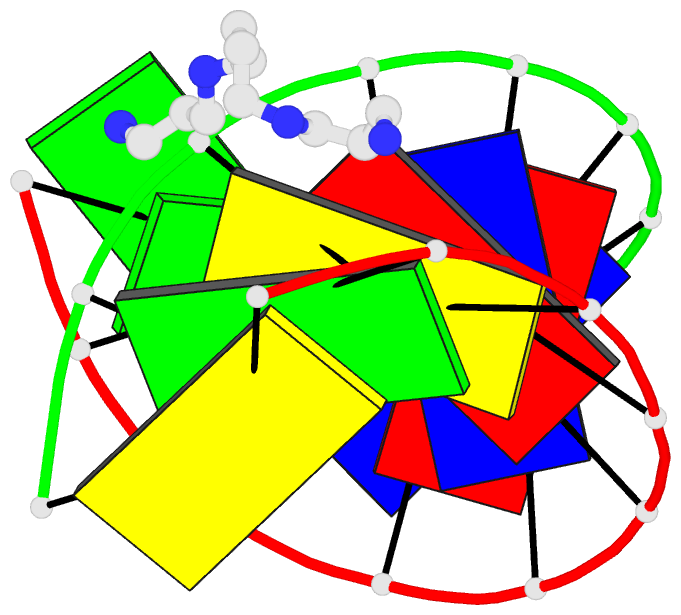
Summary information and primary citation
- PDB-id
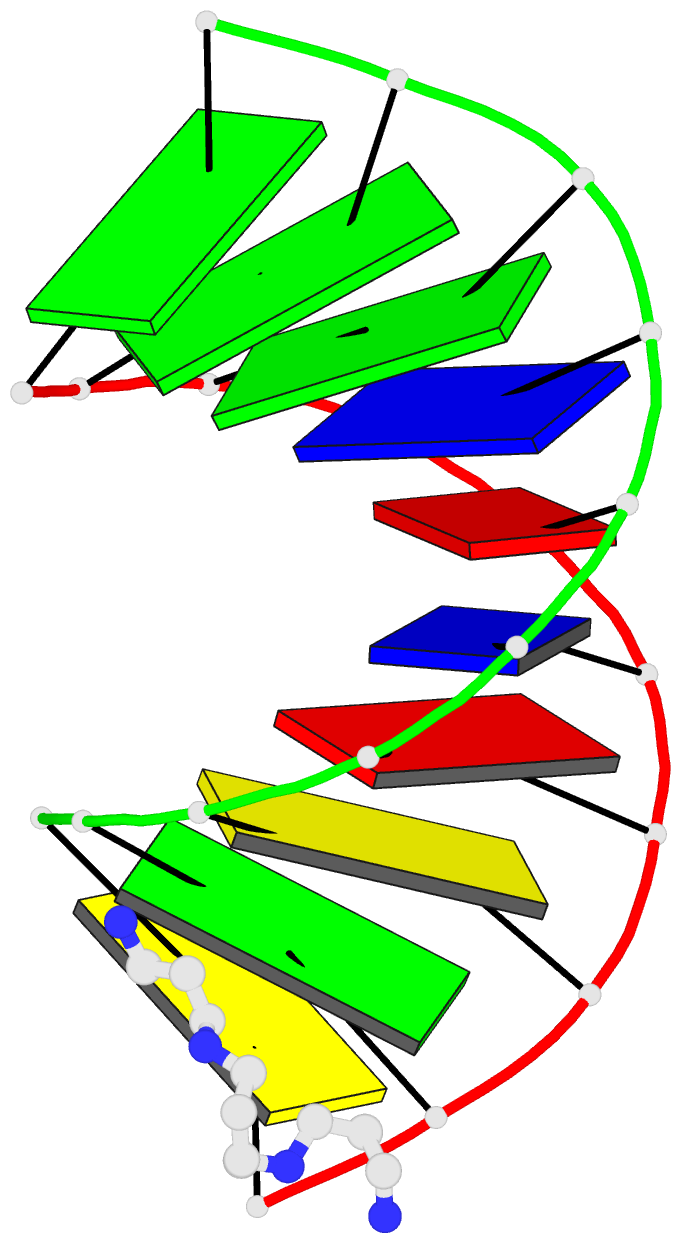
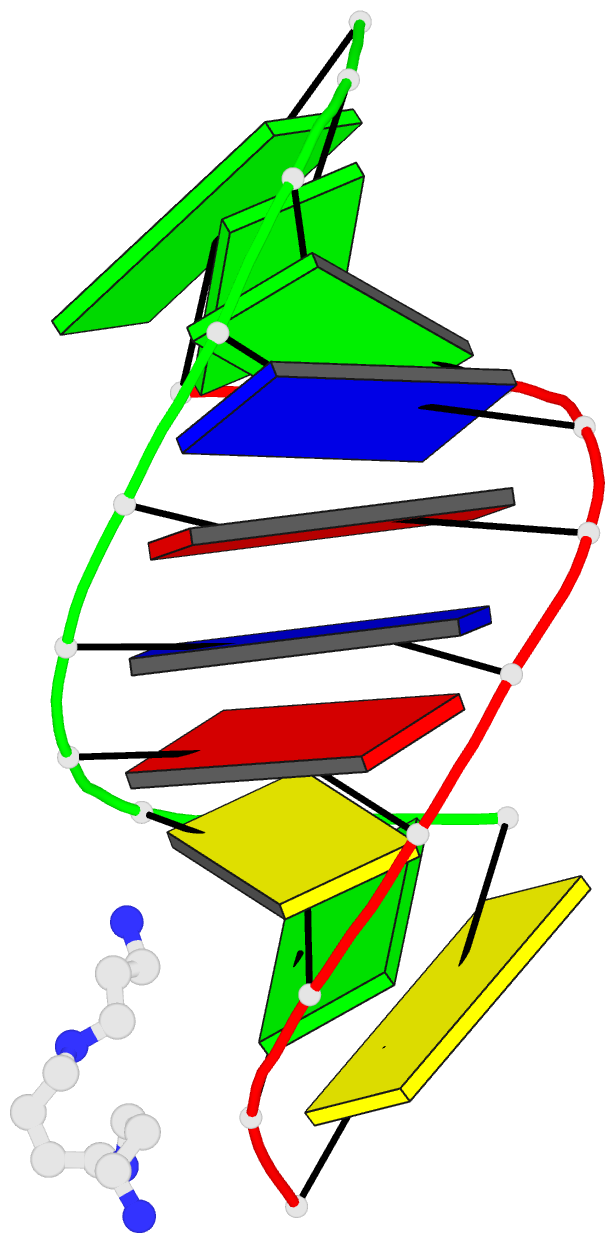
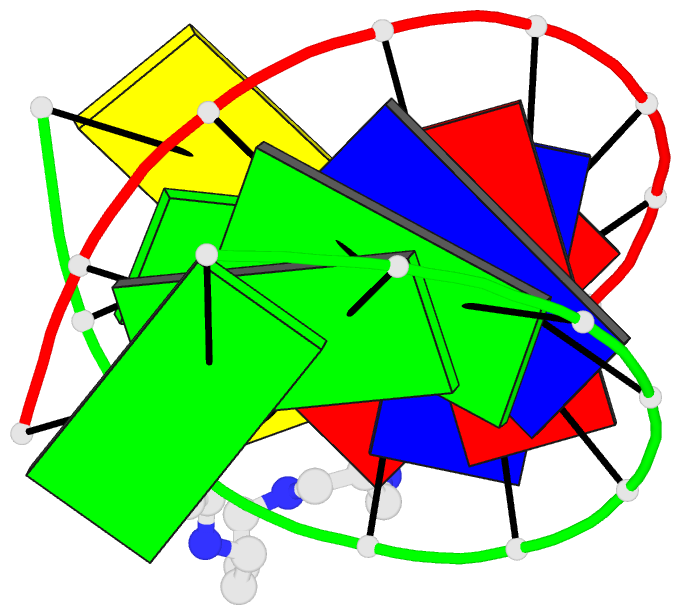
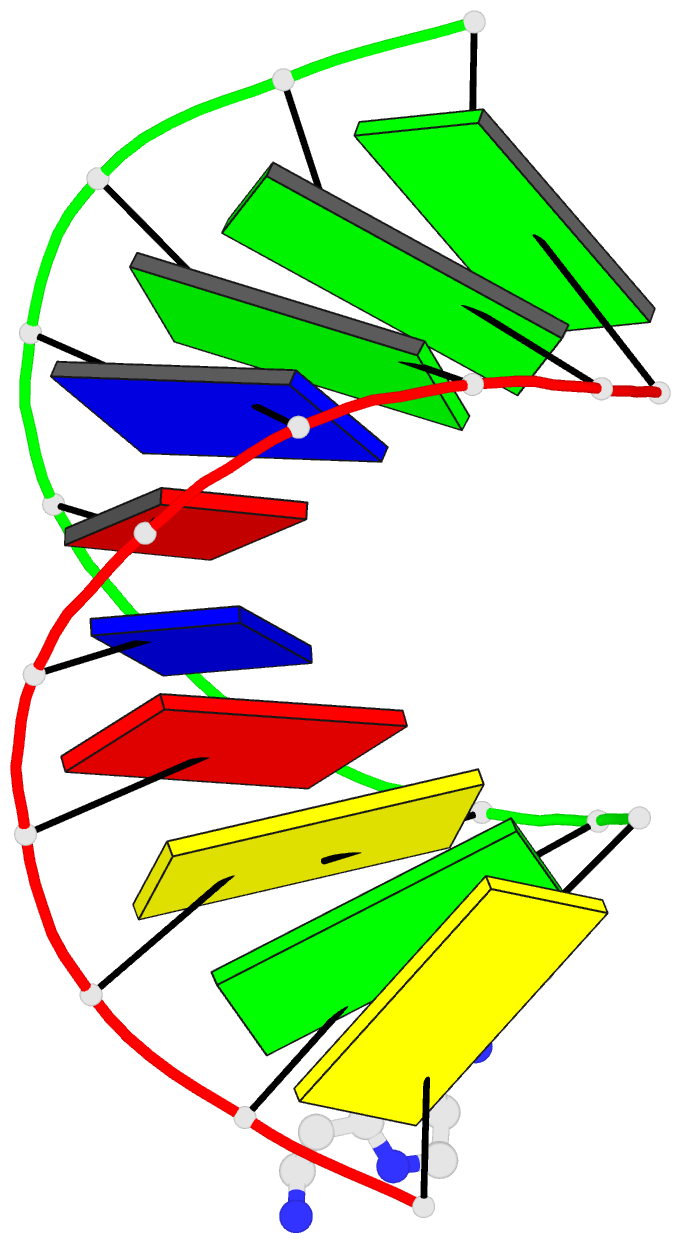
- 1ofx; DSSR-derived features in text and JSON formats
- Class
- DNA-RNA hybrid
- Method
- X-ray (2.0 Å)
- Summary
- Crystal structure of an okazaki fragment at 2 angstroms resolution
- Reference
- Egli M, Usman N, Zhang SG, Rich A (1992): "Crystal structure of an Okazaki fragment at 2-A resolution." Proc.Natl.Acad.Sci.USA, 89, 534-538. doi: 10.1073/pnas.89.2.534.
- Abstract
- In DNA replication, Okazaki fragments are formed as double-stranded intermediates during synthesis of the lagging strand. They are composed of the growing DNA strand primed by RNA and the template strand. The DNA oligonucleotide d(GGGTATACGC) and the chimeric RNA-DNA oligonucleotide r(GCG)d(TATACCC) were combined to form a synthetic Okazaki fragment and its three-dimensional structure was determined by x-ray crystallography. The fragment adopts an overall A-type conformation with 11 residues per turn. Although the base-pair geometry, particularly in the central TATA part, is distorted, there is no evidence for a transition from the A- to the B-type conformation at the junction between RNA.DNA hybrid and DNA duplex. The RNA trimer may, therefore, lock the complete fragment in an A-type conformation.