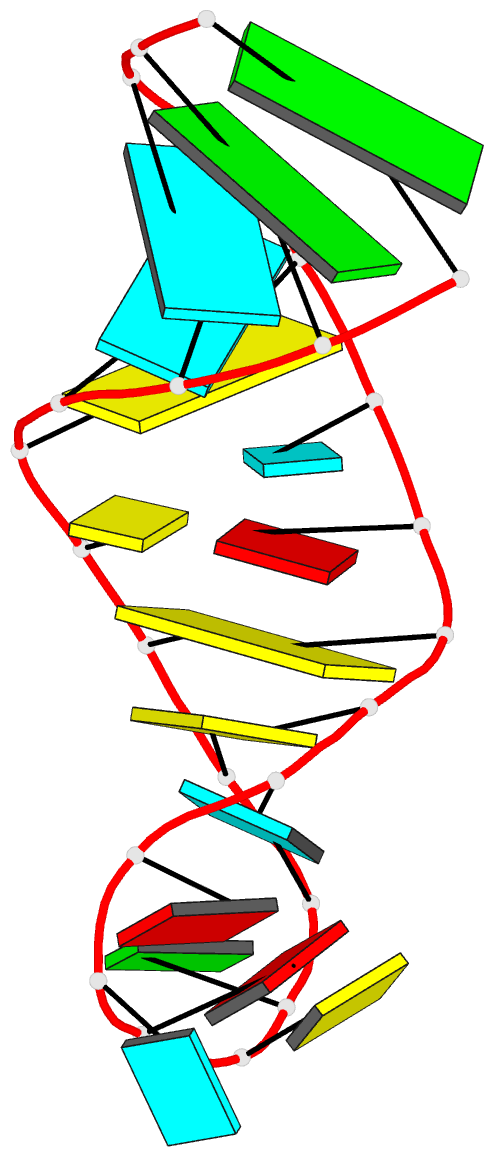
Summary information and primary citation
- PDB-id
-
1nz1;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- NMR
- Summary
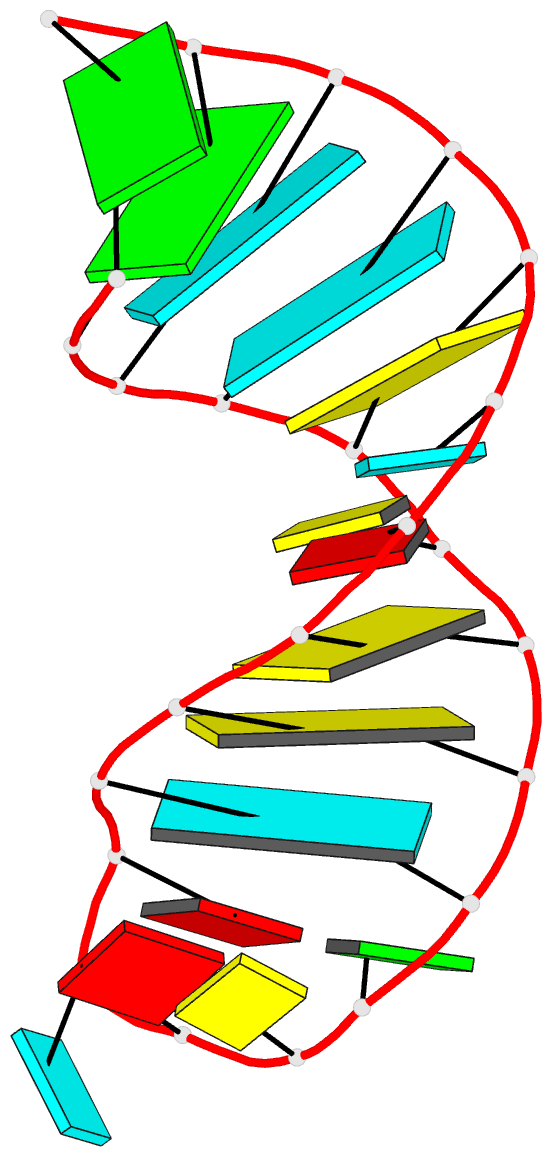
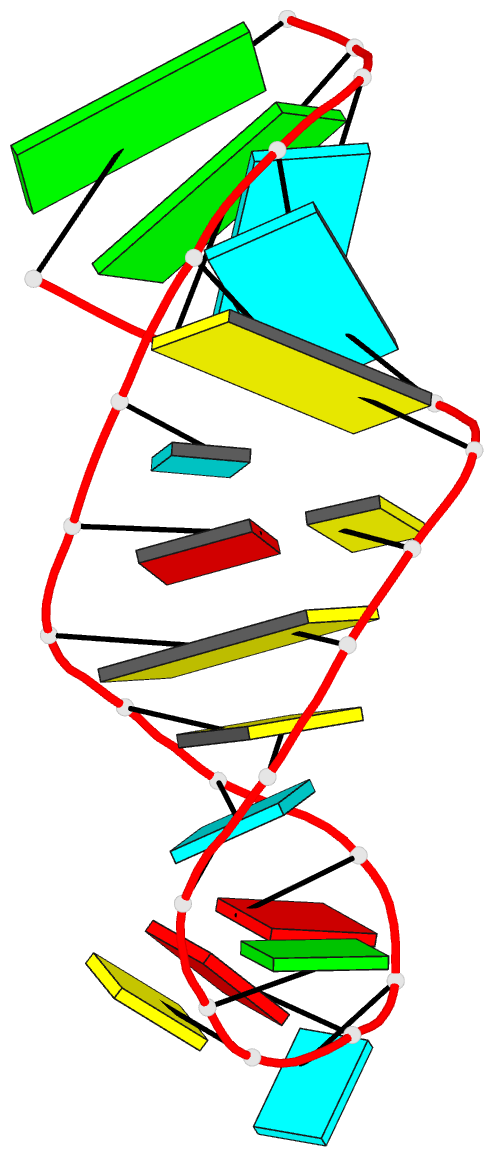
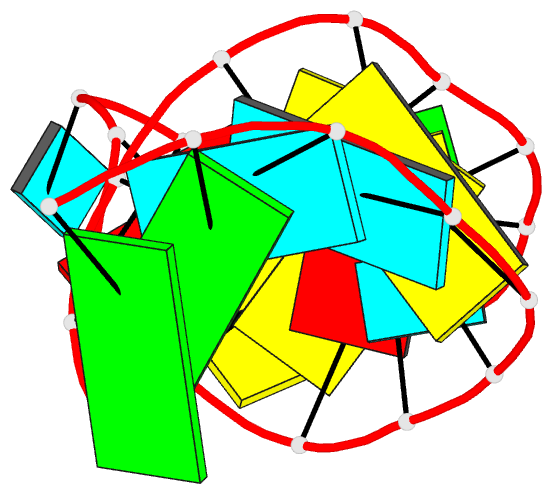
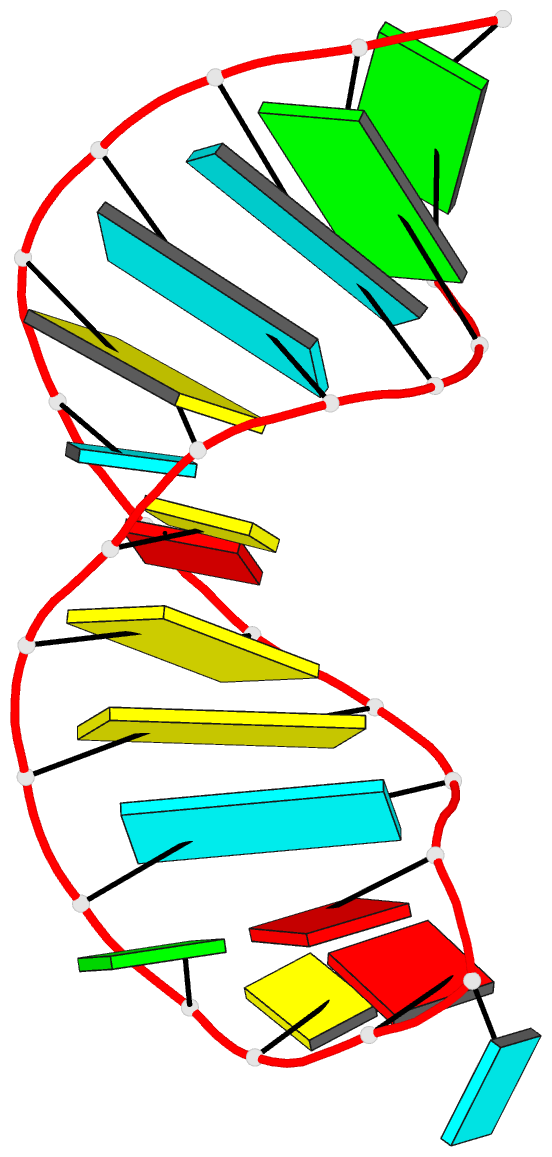
- Solution structure of the s. cerevisiae u6
intramolecular stem-loop containing an sp phosphorothioate
at nucleotide u80
- Reference
-
Reiter NJ, Nikstad LJ, Allman AM, Johnson RJ, Butcher SE
(2003): "Structure
of the U6 RNA intramolecular stem-loop harboring an
S(P)-phosphorothioate modification." RNA,
9, 533-542. doi: 10.1261/rna.2199103.
- Abstract
- Phosphorothioate-substitution experiments are often
used to elucidate functionally important metal ion-binding
sites on RNA. All previous experiments with
S(P)-phosphorothioate-substituted RNAs have been done in
the absence of structural information for this particular
diastereomer. Yeast U6 RNA contains a metal ion-binding
site that is essential for spliceosome function and
includes the pro-S(P) oxygen 5' of U(80).
S(P)-phosphorothioate substitution at this location creates
spliceosomes dependent on thiophilic ions for the first
step of splicing. We have determined the solution structure
of the U(80) S(P)-phosphorothioate-substituted U6
intramolecular stem-loop (ISL), and also report the refined
NMR structure of the unmodified U6 ISL. Both structures
were determined with inclusion of (1)H-(13)C residual
dipolar couplings. The precision of the structures with and
without phosphorothioate (RMSD = 1.05 and 0.79 A,
respectively) allows comparison of the local and long-range
structural effect of the modification. We find that the
U6-ISL structure is unperturbed by the phosphorothioate.
Additionally, the thermodynamic stability of the U6 ISL is
dependent on the protonation state of the A(79)-C(67)
wobble pair and is not affected by the adjacent
phosphorothioate. These results indicate that a single
S(P)-phosphorothioate substitution can be structurally
benign, and further validate the metal ion rescue
experiments used to identify the essential metal-binding
site(s) in the spliceosome.