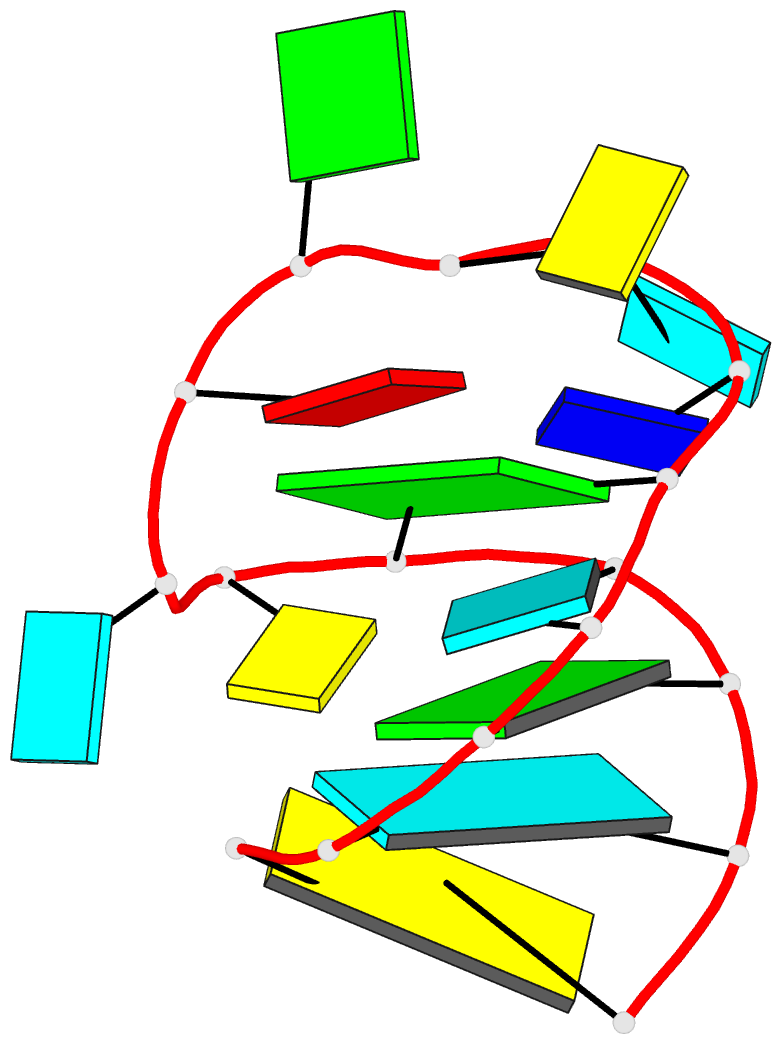
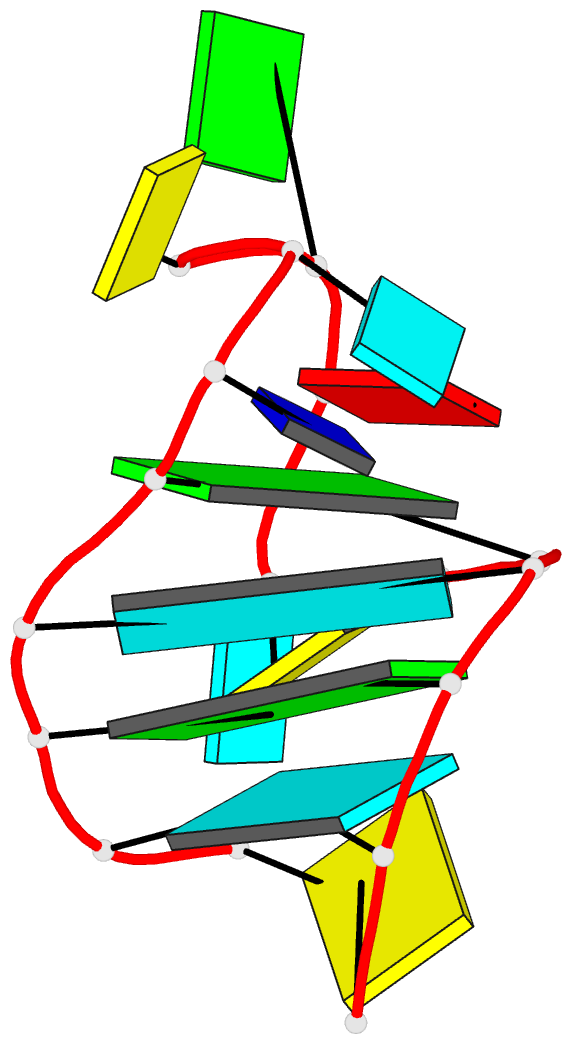
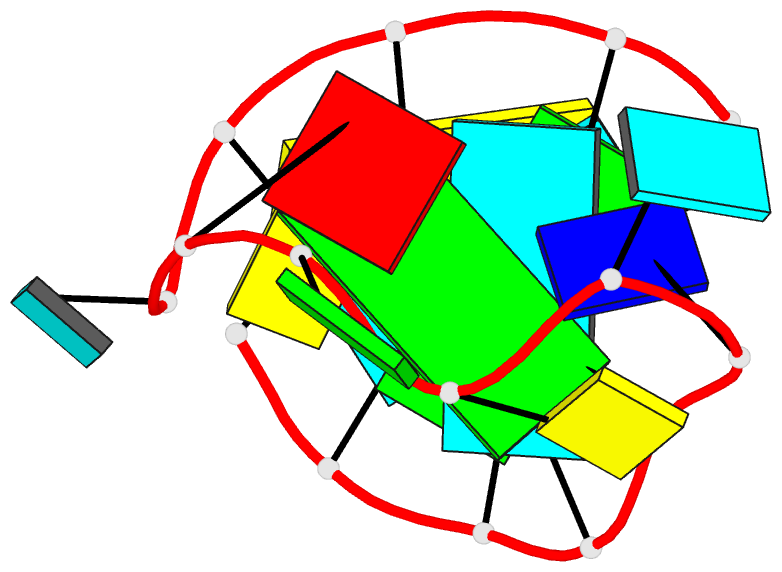
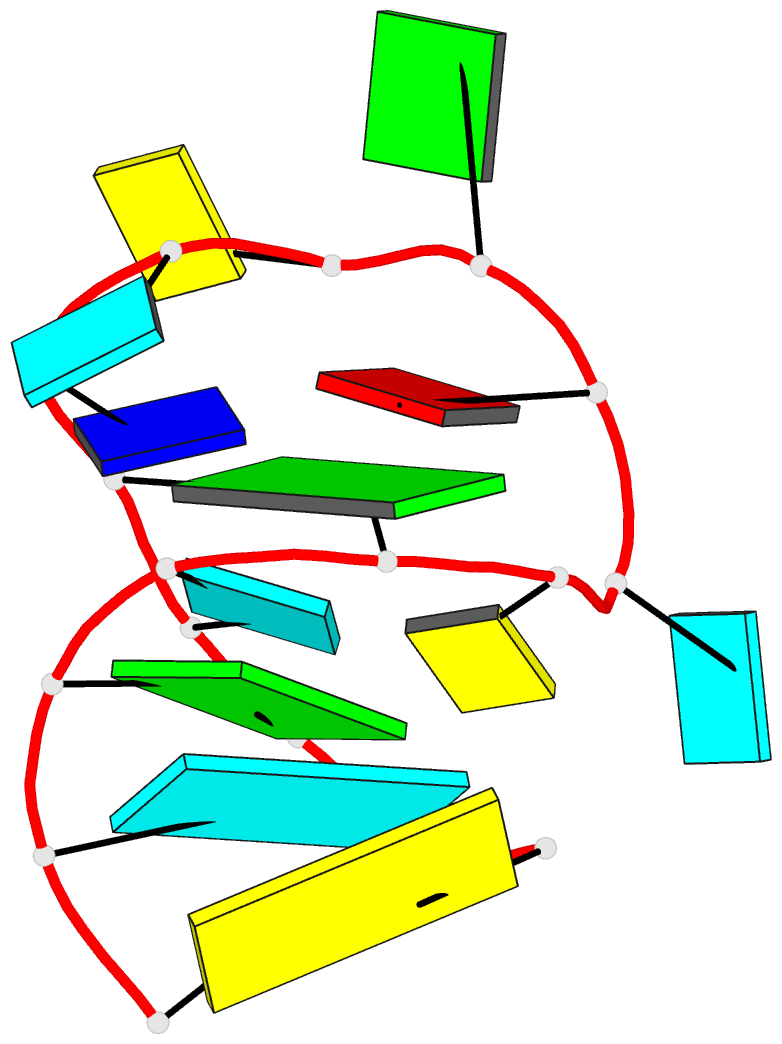
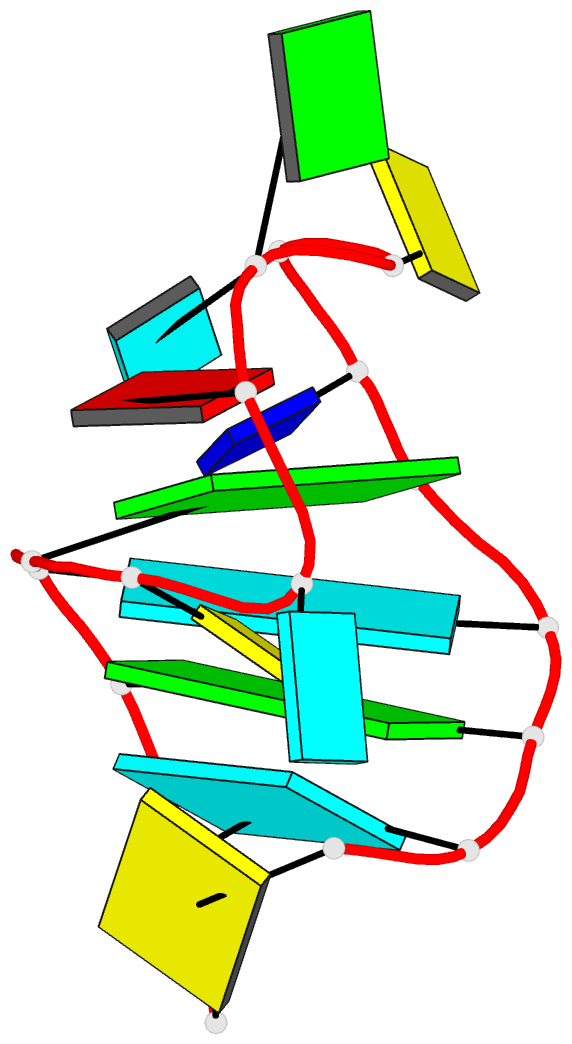
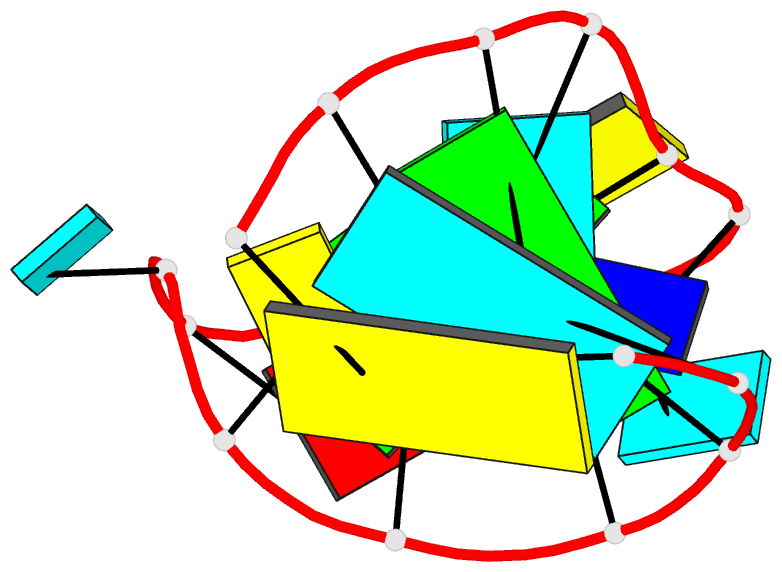
Summary information and primary citation
- PDB-id
- 1kos; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Solution NMR structure of an analog of the yeast trna phe t stem loop containing ribothymidine at its naturally occurring position
- Reference
- Koshlap KM, Guenther R, Sochacka E, Malkiewicz A, Agris PF (1999): "A distinctive RNA fold: the solution structure of an analogue of the yeast tRNAPhe T Psi C domain." Biochemistry, 38, 8647-8656. doi: 10.1021/bi990118w.
- Abstract
- The structure of an analogue of the yeast tRNAPhe T Psi C stem-loop has been determined by NMR spectroscopy and restrained molecular dynamics. The molecule contained the highly conserved modification ribothymidine at its naturally occurring position. The ribothymidine-modified T Psi C stem-loop is the product of the m5U54-tRNA methyltransferase, but is not a substrate for the m1A58-tRNA methyltransferase. Site-specific substitutions and 15N labels were used to confirm the assignment of NOESY cross-peaks critical in defining the global fold of the molecule. The structure is unusual in that the loop folds far over into the major groove of the curved stem. This conformation is stabilized by both stacking interactions and hydrogen bond formation. Furthermore, this conformation appears to be unique among RNA hairpins of similar size. There is, however, a considerable resemblance to the analogous domain in the crystal structure of the full-length yeast tRNAPhe. We believe, therefore, that the structure we have determined may represent an intermediate in the folding pathway during the maturation of tRNA.