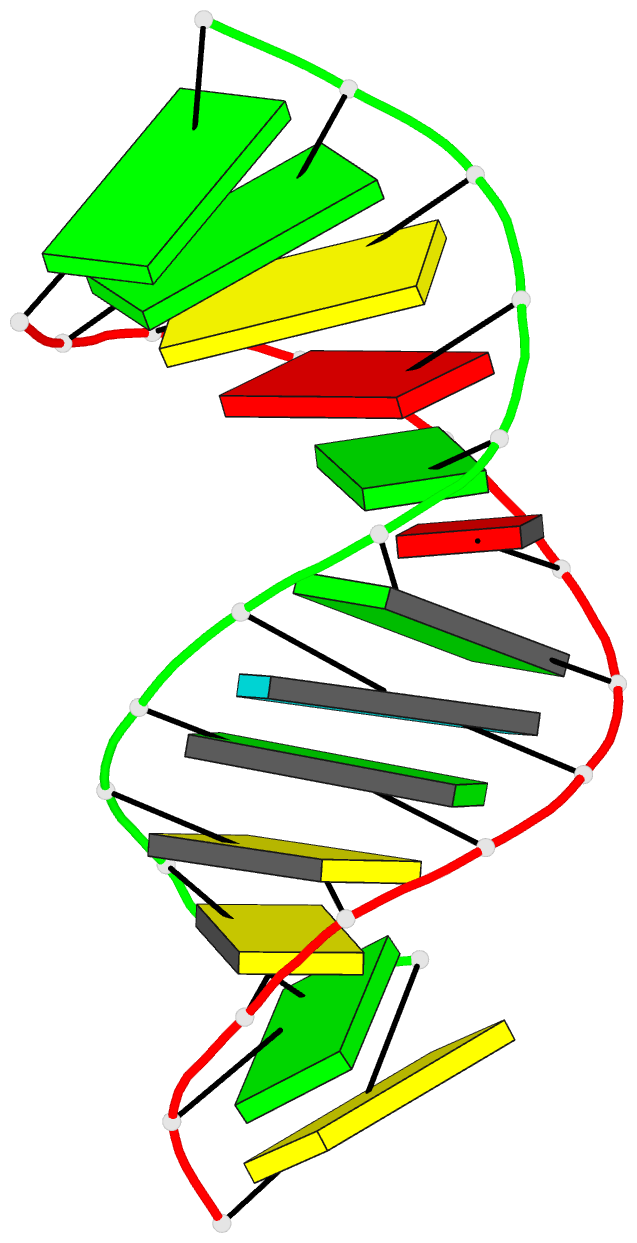
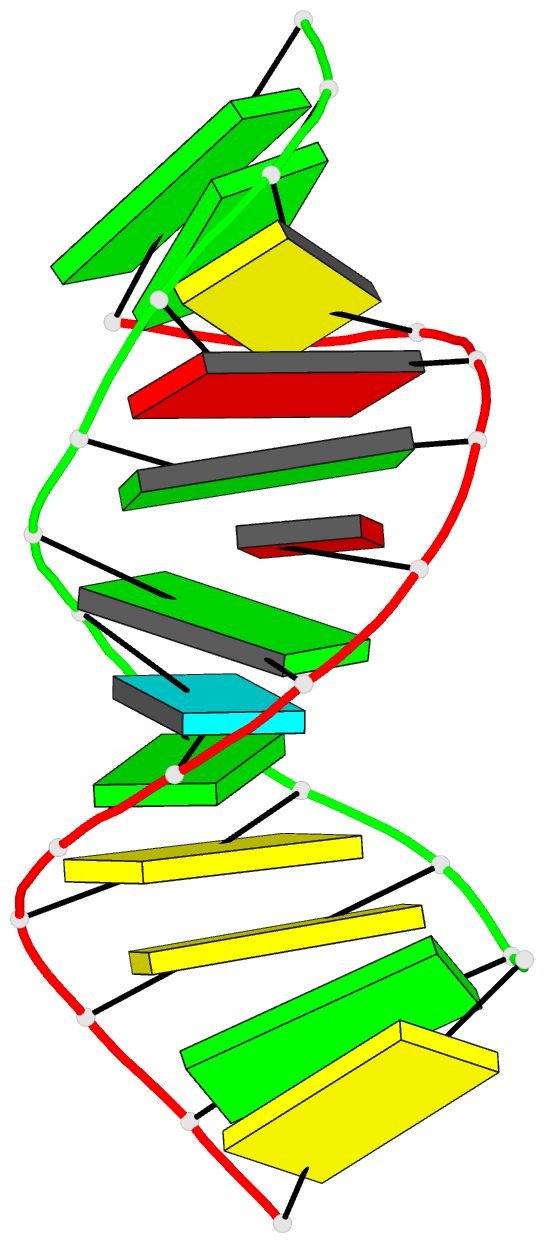
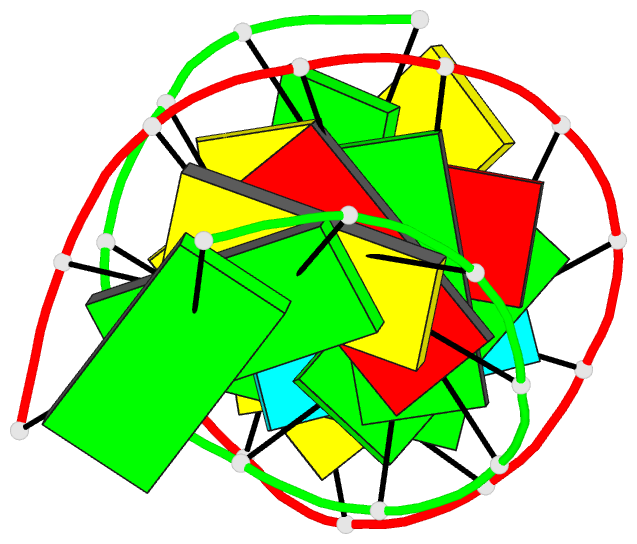
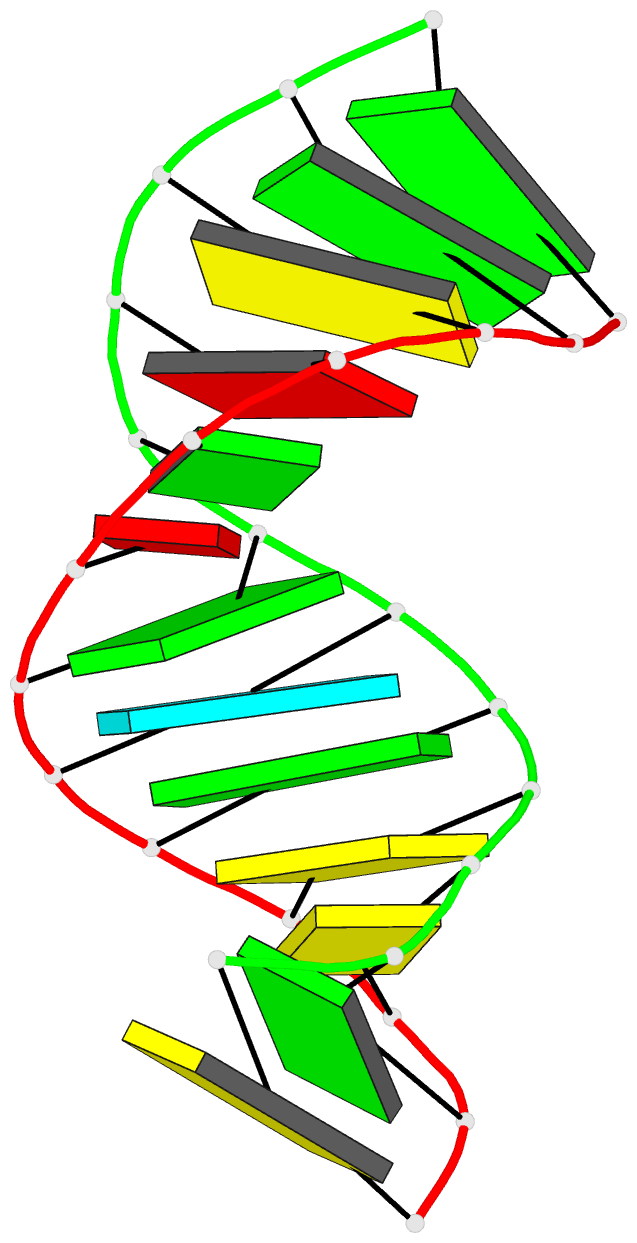
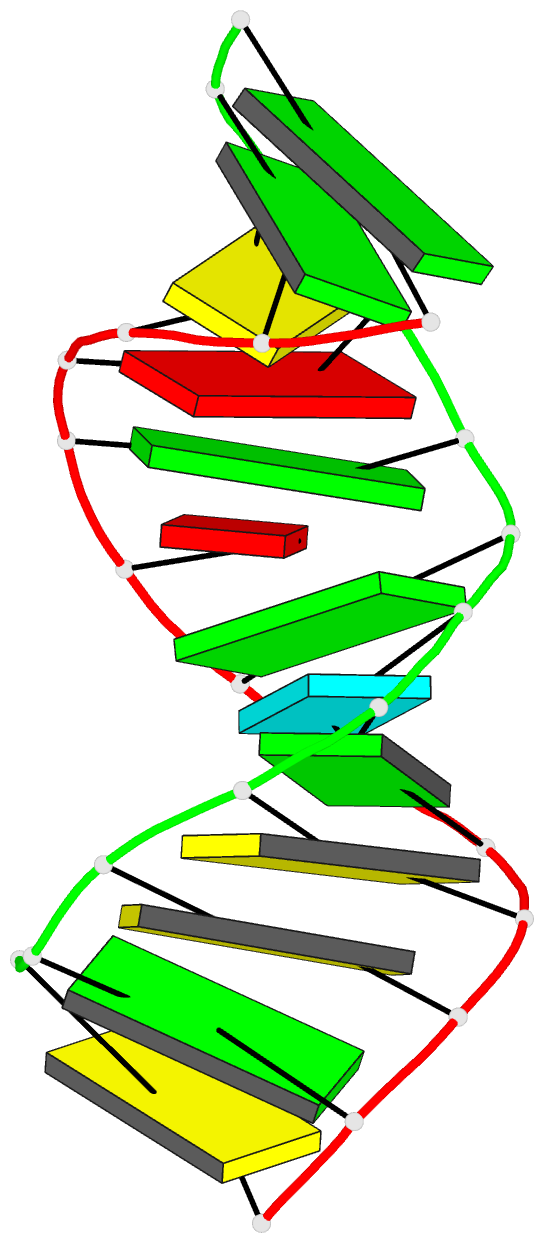
Summary information and primary citation
- PDB-id
-
1k8s;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Bulged adenosine in an RNA duplex
- Reference
-
Thiviyanathan V, Guliaev AB, Leontis NB, Gorenstein DG
(2000): "Solution
conformation of a bulged adenosine base in an RNA duplex
by relaxation matrix refinement."
J.Mol.Biol., 300, 1143-1154.
doi: 10.1006/jmbi.2000.3931.
- Abstract
- Bulges are common structural motifs in RNA secondary
structure and are thought to play important roles in
RNA-protein and RNA-drug interactions. Adenosine bases are
the most commonly occurring unpaired base in double helical
RNA secondary structures. The solution conformation and
dynamics of a 25-nucleotide RNA duplex containing an
unpaired adenosine, r(GGCAGAGUGCCGC): r(GCGGCACCUGCC) have
been studied by NMR spectroscopy and MORASS iterative
relaxation matrix structural refinement. The results show
that the bulged adenosine residue stacks into the RNA
duplex with little perturbation around the bulged region.
Most of the bases in the RNA duplex adopt C(3)'-endo
conformation, exhibiting the N-type sugar pucker as found
in the A form helices. The sugars of the bulged residue and
the 5' flanking residue to it are found to exhibit
C(2)'-endo conformation. None of the residues are in syn
conformation.