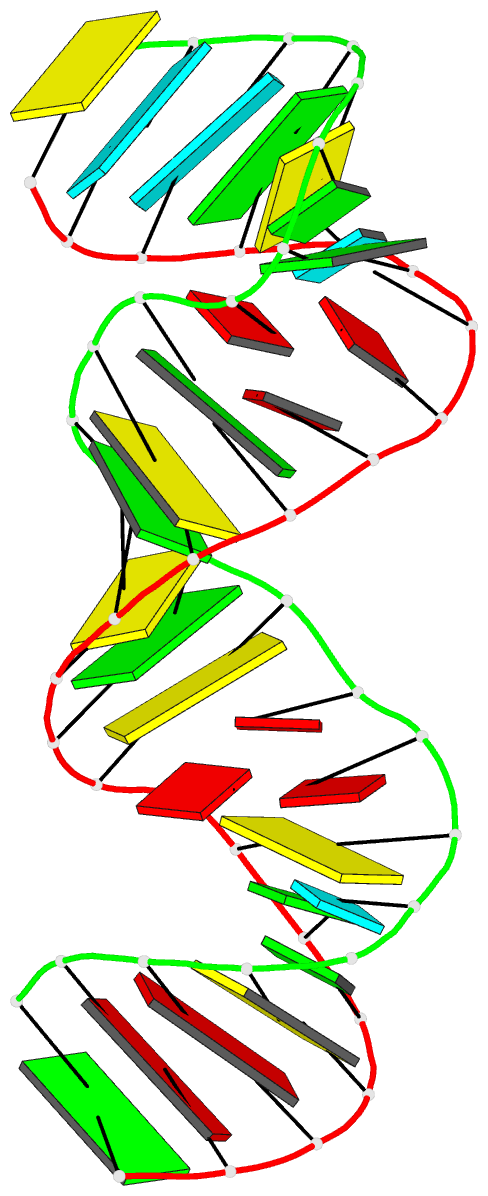
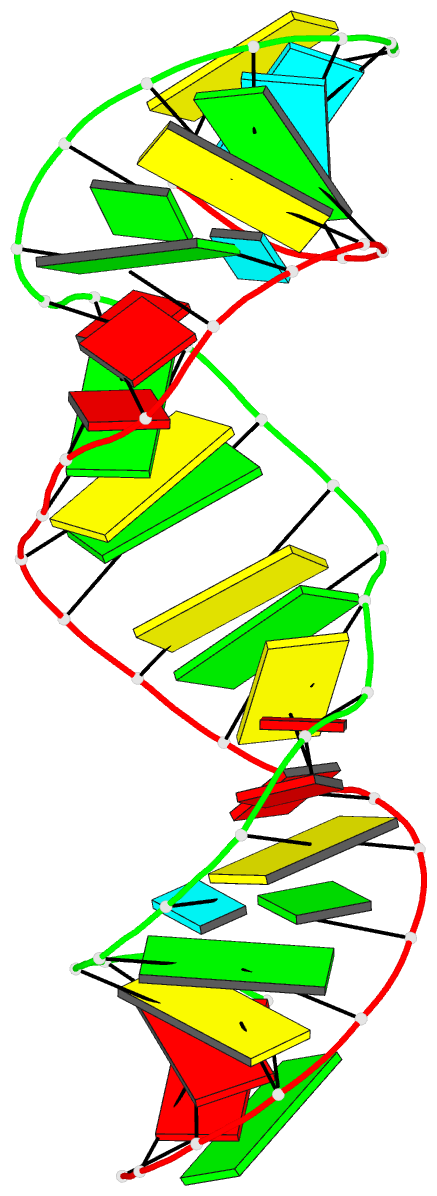
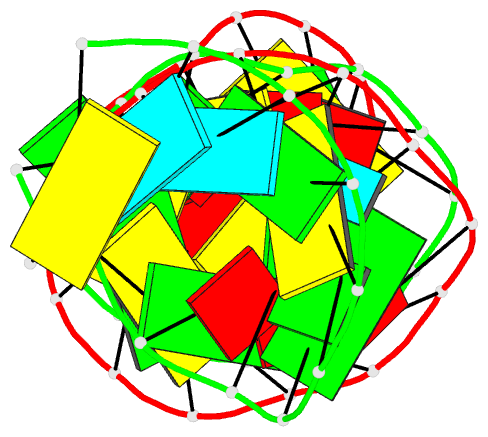
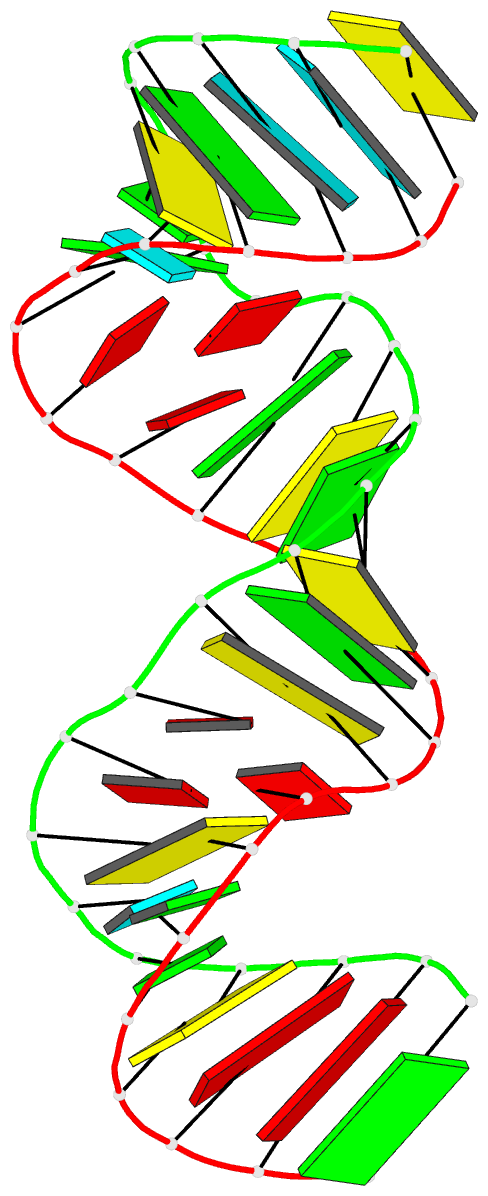
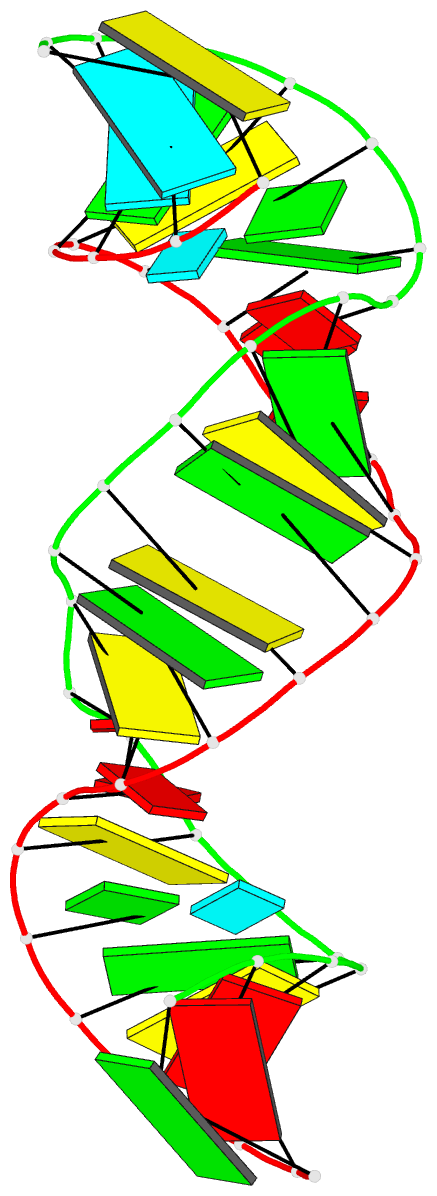
Summary information and primary citation
- PDB-id
- 1ju1; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Dimer initiation sequence of hiv-1lai genomic RNA: NMR solution structure of the extended duplex
- Reference
- Girard F, Barbault F, Gouyette C, Huynh-Dinh T, Paoletti J, Lancelot G (1999): "Dimer initiation sequence of HIV-1Lai genomic RNA: NMR solution structure of the extended duplex." J.Biomol.Struct.Dyn., 16, 1145-1157.
- Abstract
- The genome of all retrovirus consists of two copies of genomic RNA which are noncovalently linked near their 5' end. A sequence localized immediately upstream from the splice donor site inside the HIV-1 psi-RNA region was identified as the domain responsible for the dimerization initiation. It was shown that a kissing complex and a stable dimer are both involved in the HIV-1Lai RNA dimerization process in vitro. The NCp7 protein activates the dimerization by converting a transient loop-loop complex into a more stable dimer. The structure of this transitory loop-loop complex was recently elucidated by Mujeeb et al. In work presented here, we use NMR spectroscopy to determine the stable extended dimer structure formed from a 23 nucleotides RNA fragment, part of the 35 nucleotides SL1 sequence. By heating to 90 degrees C, then slowly cooling this sequence, we were able to show that an extended dimer is formed. We present evidence for the three dimensional structure of this dimer. NMR data yields evidence for a zipper like motif A8A9.A16 existence. This motif enables the surrounding bases to be positioned more closely and permit the G7 and C17 bases to be paired. This is different to other related sequences where only the kissing complex is observed, we suggest that the zipper like motif AA.A could be an important stabilization factor of the extended duplex.