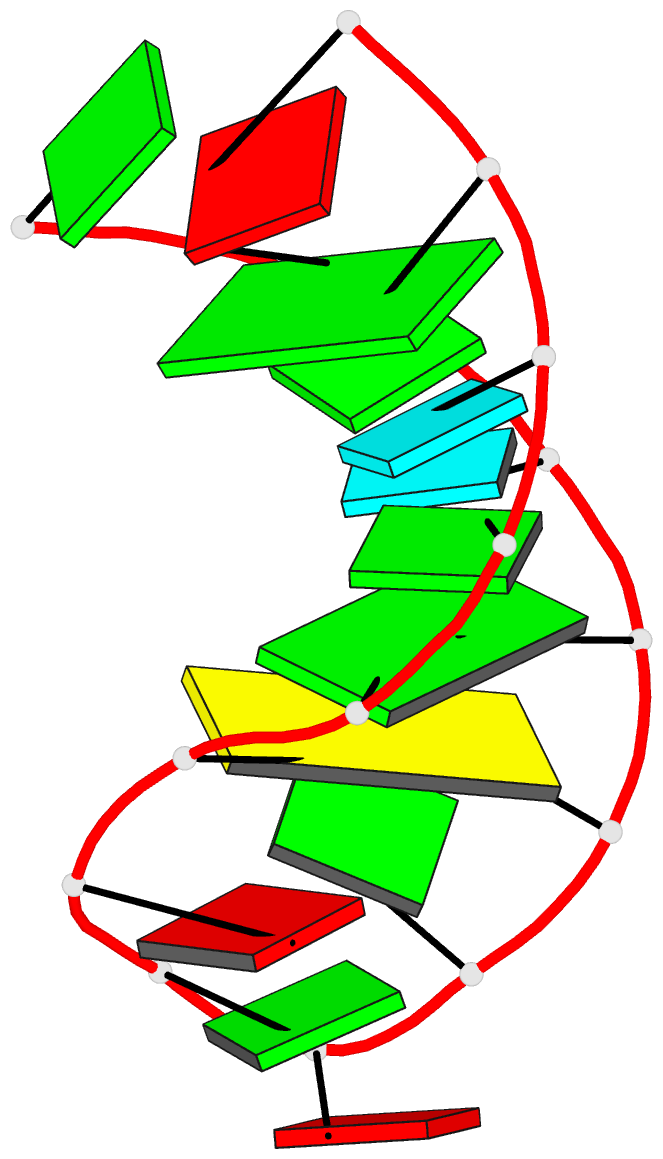
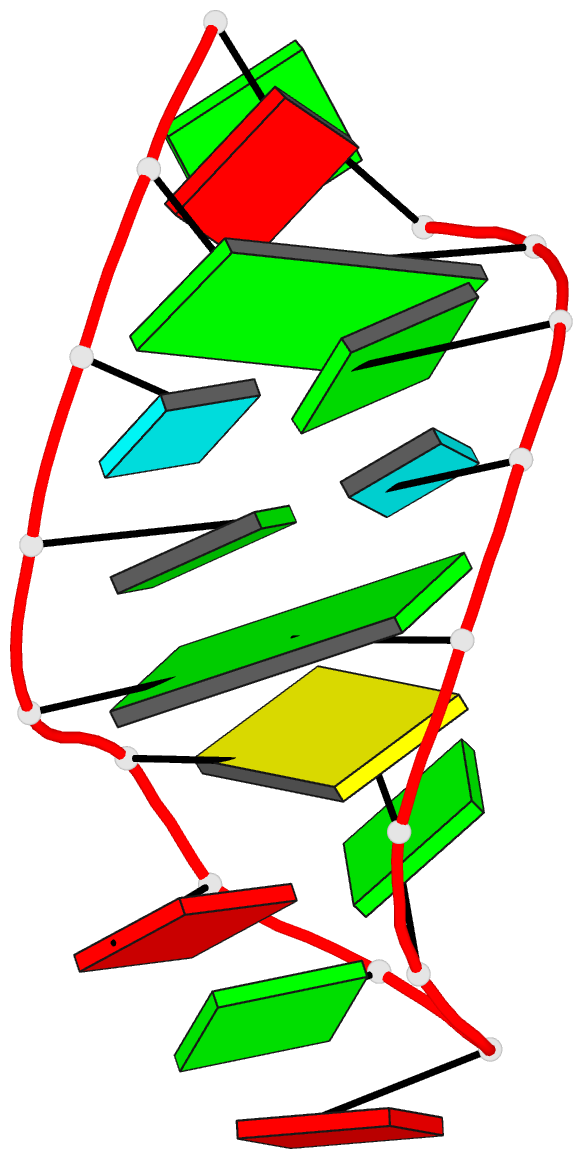
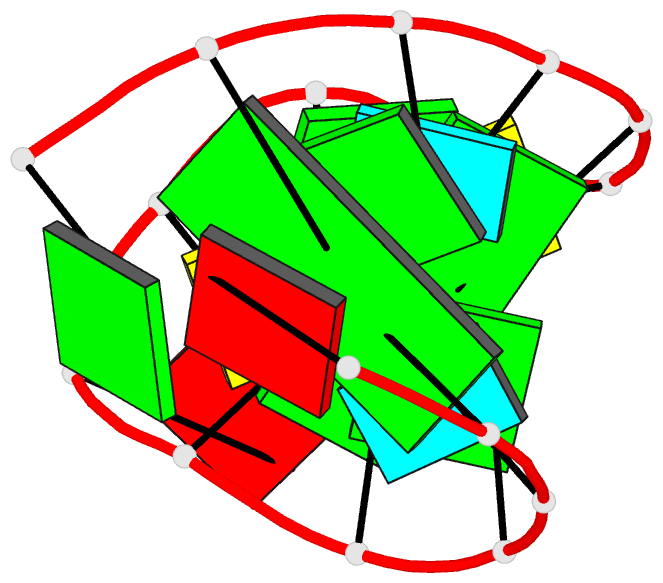
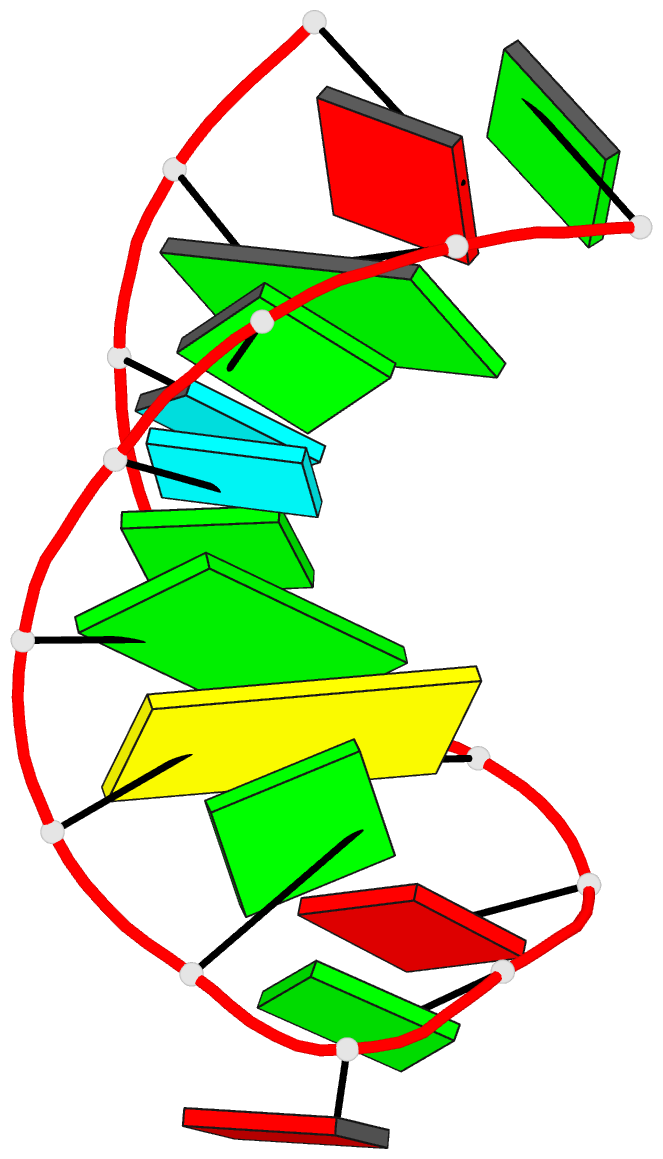
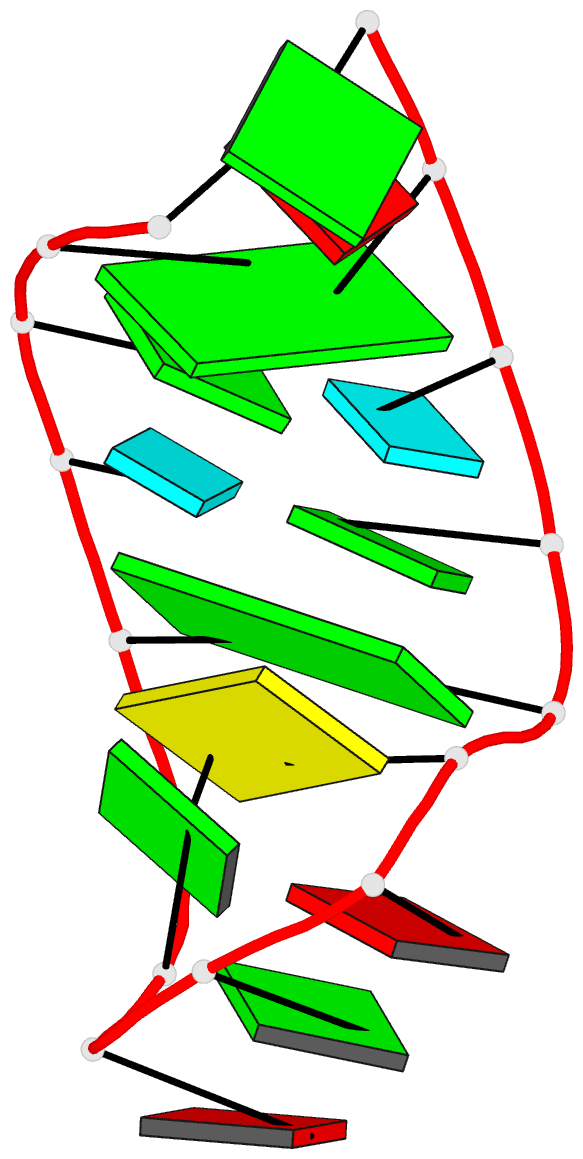
Summary information and primary citation
- PDB-id
- 1jtw; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Structure of sl4 RNA from the hiv-1 packaging signal
- Reference
- Kerwood DJ, Cavaluzzi MJ, Borer PN (2001): "Structure of SL4 RNA from the HIV-1 packaging signal." Biochemistry, 40, 14518-14529. doi: 10.1021/bi0111909.
- Abstract
- The NMR-based structure is described for an RNA model of stem-loop 4 (SL4) from the HIV-1 major packaging domain. The GAGA tetraloop adopts a conformation similar to the classic GNRA form, although there are differences in the details. The type II tandem G.U pairs have a combination of wobble and bifurcated hydrogen bonds where the uracil 2-carbonyl oxygen is hydrogen-bonded to both G,H1 and G,H2. There is the likelihood of a Na(+) ion coordinated to the four carbonyl oxygens in the major groove for these G.U pairs and perhaps to the N7 lone pairs of the G bases as well. A continuous stack of five bases extends over nearly the whole length of the stem to the base of the loop in the RNA 16mer: C15/U14/G13/G5/C6. There is no evidence for a terminal G.A pair; instead, G1 appears quite unrestrained, and A16 stacks on both C15 and G2. Residues G2 through G5 exhibit broadened resonances, especially G3 and U4, suggesting enhanced mobility for the 5'-side of the stem. The structure shows G2/G3/U4 stacking along the same strand, nearly isolated from interaction with the other bases. This is probably an important factor in the signal broadening and apparent mobility of these residues and the low stability of the 16mer hairpin against thermal denaturation.