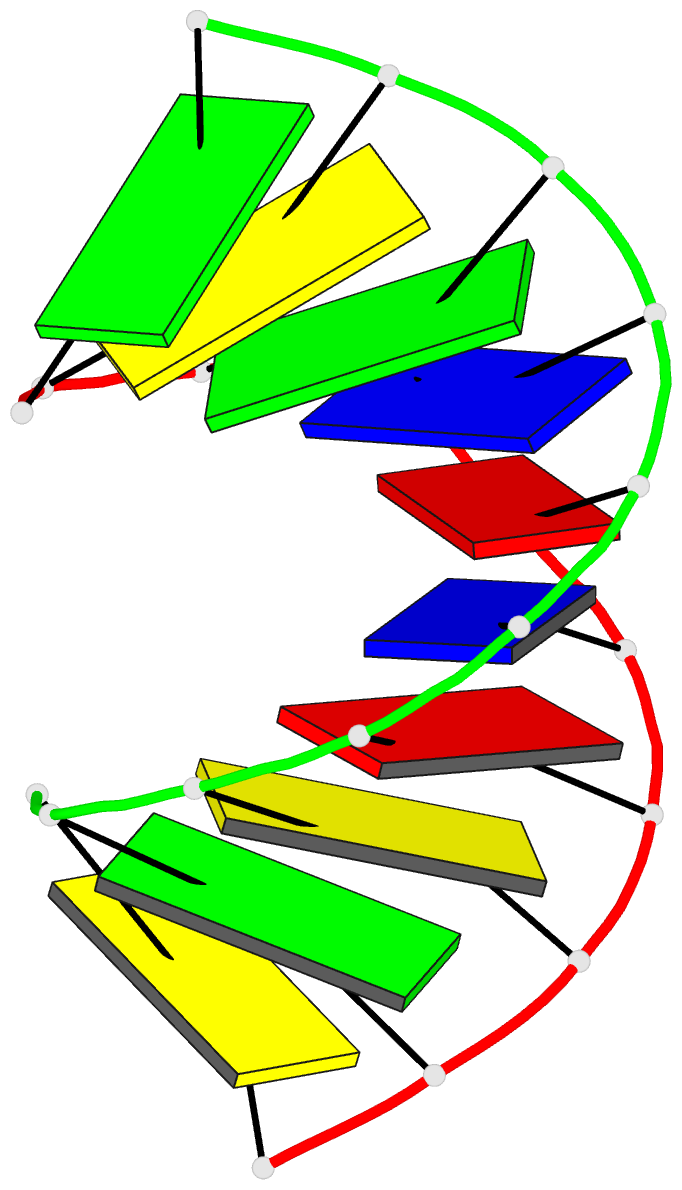
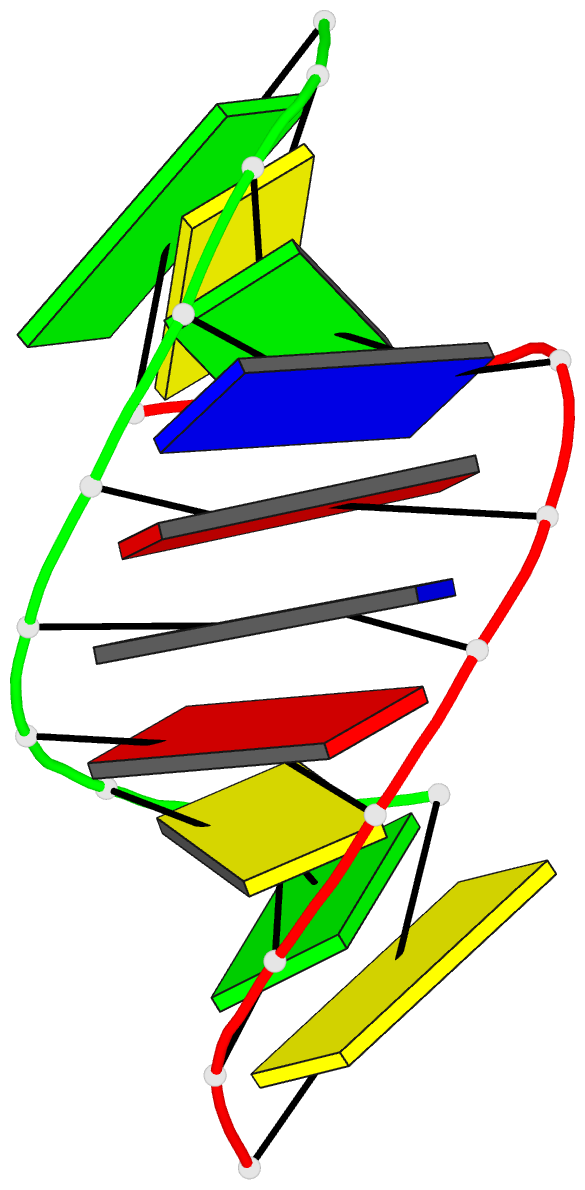
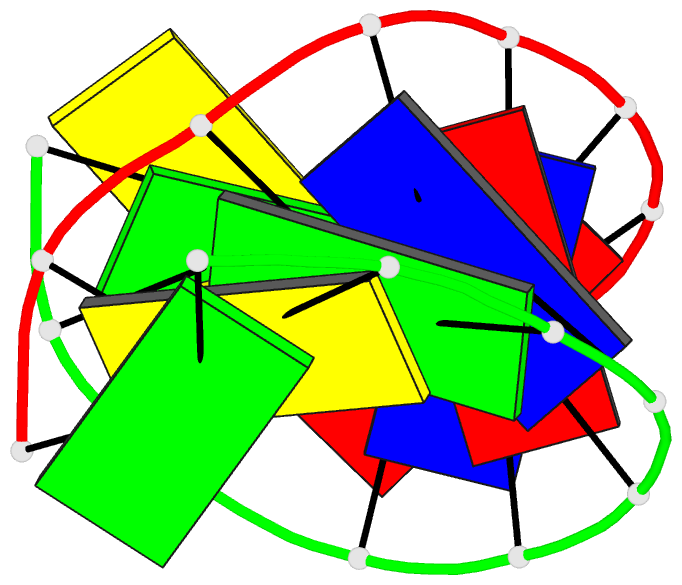
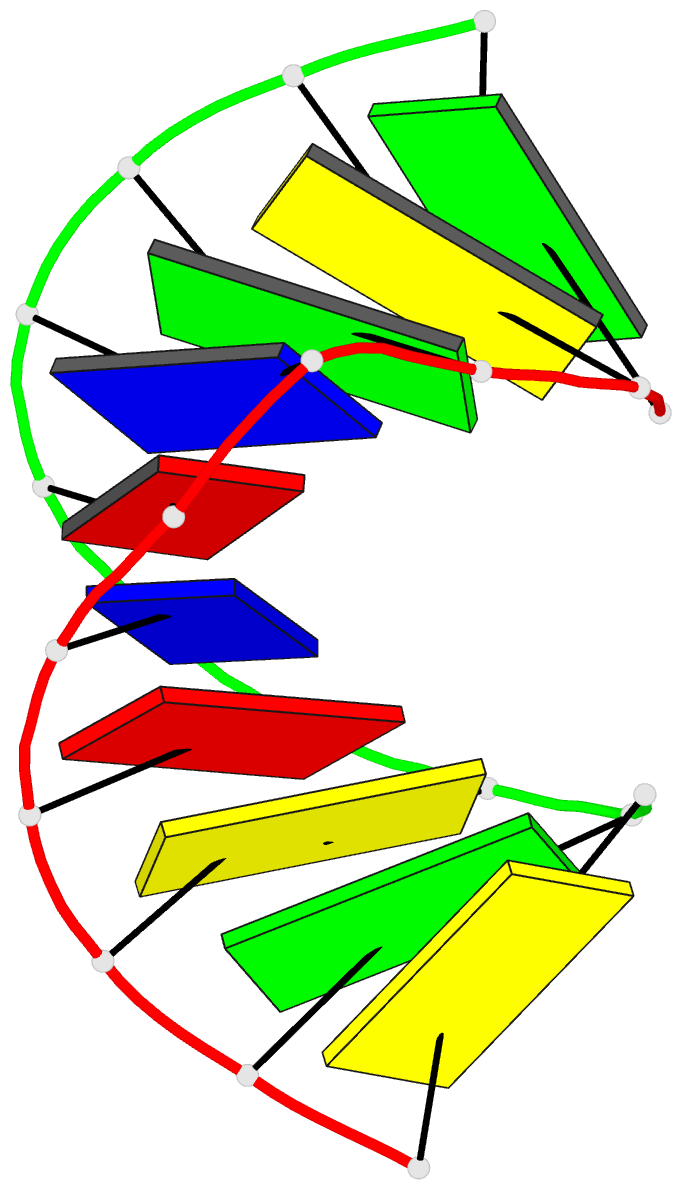
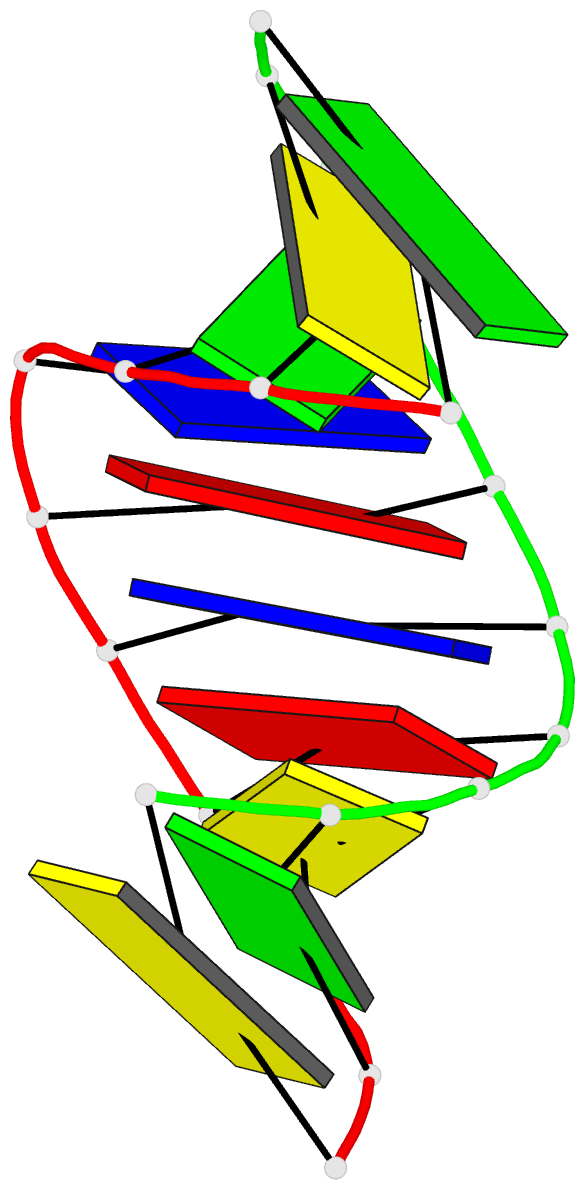
Summary information and primary citation
- PDB-id
- 1i0p; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- X-ray (1.3 Å)
- Summary
- 1.3 a structure of the a-decamer gcgtatacgc with a single 2'-o-methyl-[tri(oxyethyl)], medium k-salt
- Reference
- Tereshko V, Wilds CJ, Minasov G, Prakash TP, Maier MA, Howard A, Wawrzak Z, Manoharan M, Egli M (2001): "Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments." Nucleic Acids Res., 29, 1208-1215. doi: 10.1093/nar/29.5.1208.
- Abstract
- The observation of light metal ions in nucleic acids crystals is generally a fortuitous event. Sodium ions in particular are notoriously difficult to detect because their X-ray scattering contributions are virtually identical to those of water and Na(+.)O distances are only slightly shorter than strong hydrogen bonds between well-ordered water molecules. We demonstrate here that replacement of Na(+) by K(+), Rb(+) or Cs(+) and precise measurements of anomalous differences in intensities provide a particularly sensitive method for detecting alkali metal ion-binding sites in nucleic acid crystals. Not only can alkali metal ions be readily located in such structures, but the presence of Rb(+) or Cs(+) also allows structure determination by the single wavelength anomalous diffraction technique. Besides allowing identification of high occupancy binding sites, the combination of high resolution and anomalous diffraction data established here can also pinpoint binding sites that feature only partial occupancy. Conversely, high resolution of the data alone does not necessarily allow differentiation between water and partially ordered metal ions, as demonstrated with the crystal structure of a DNA duplex determined to a resolution of 0.6 A.