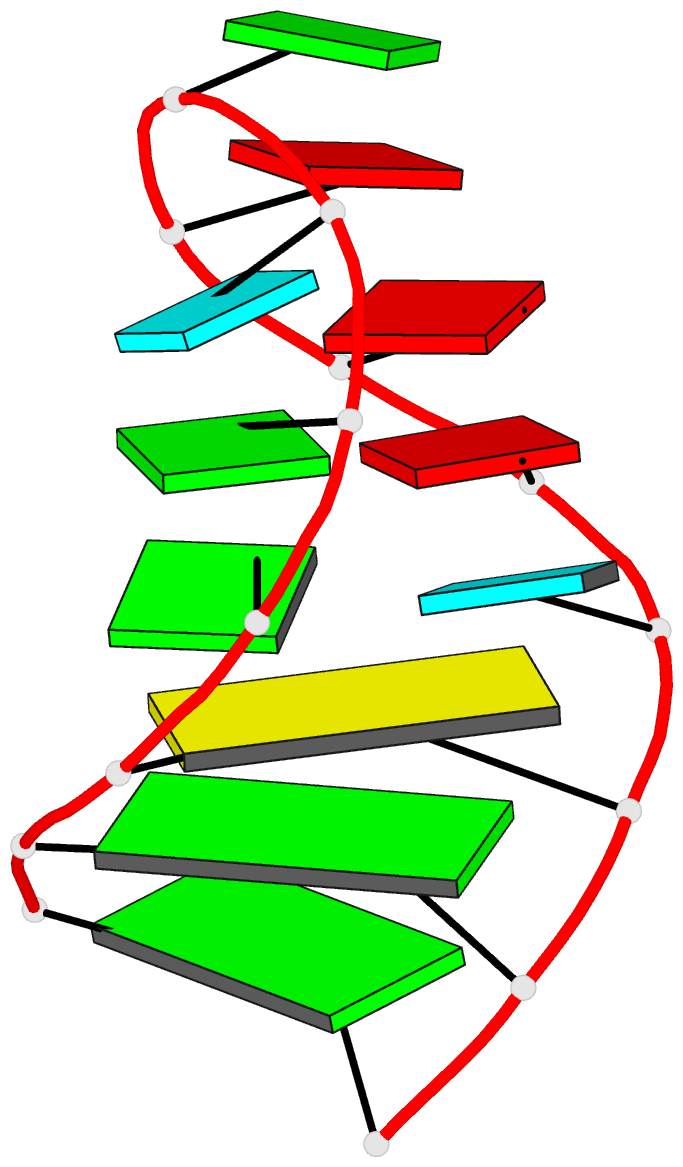
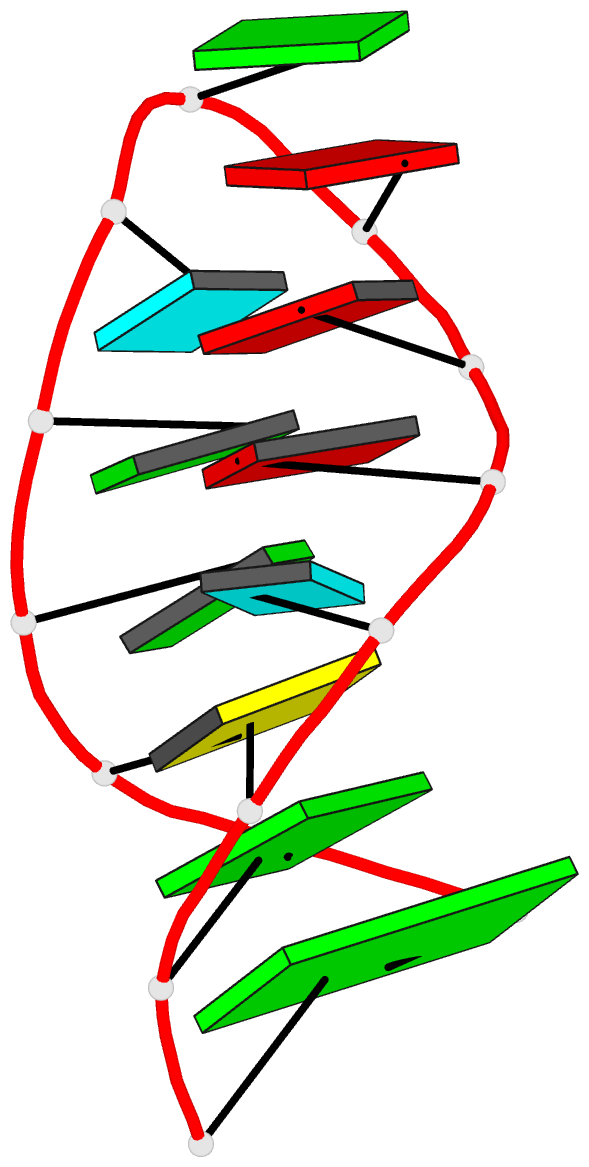
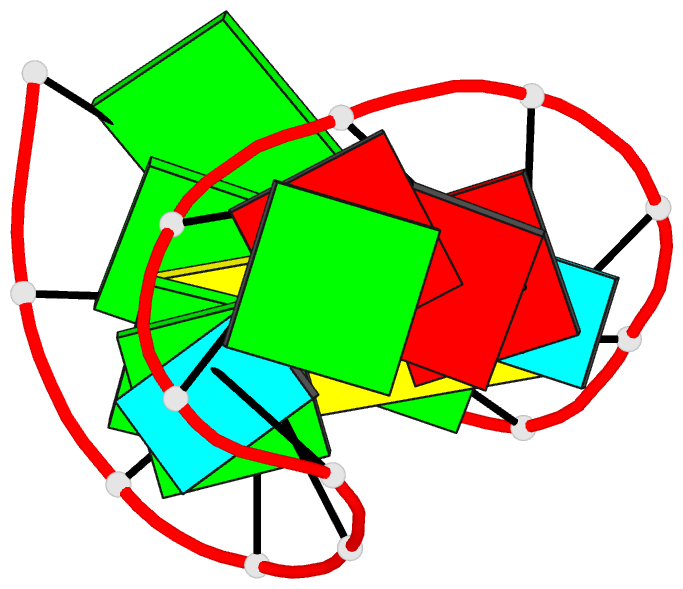
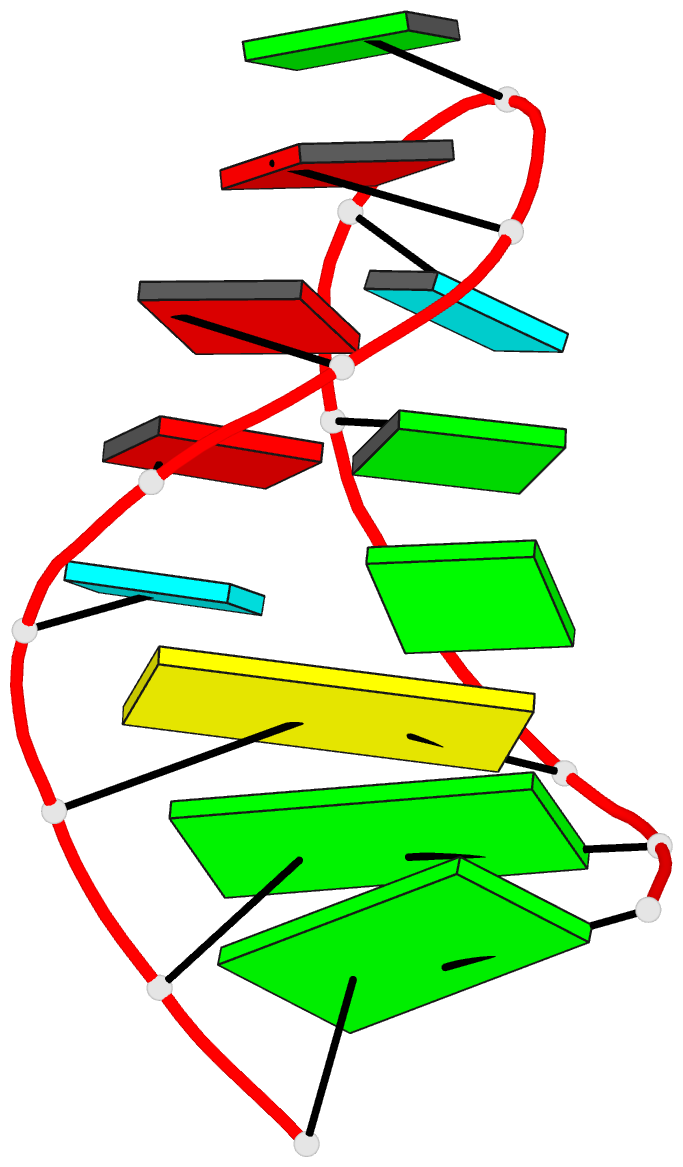
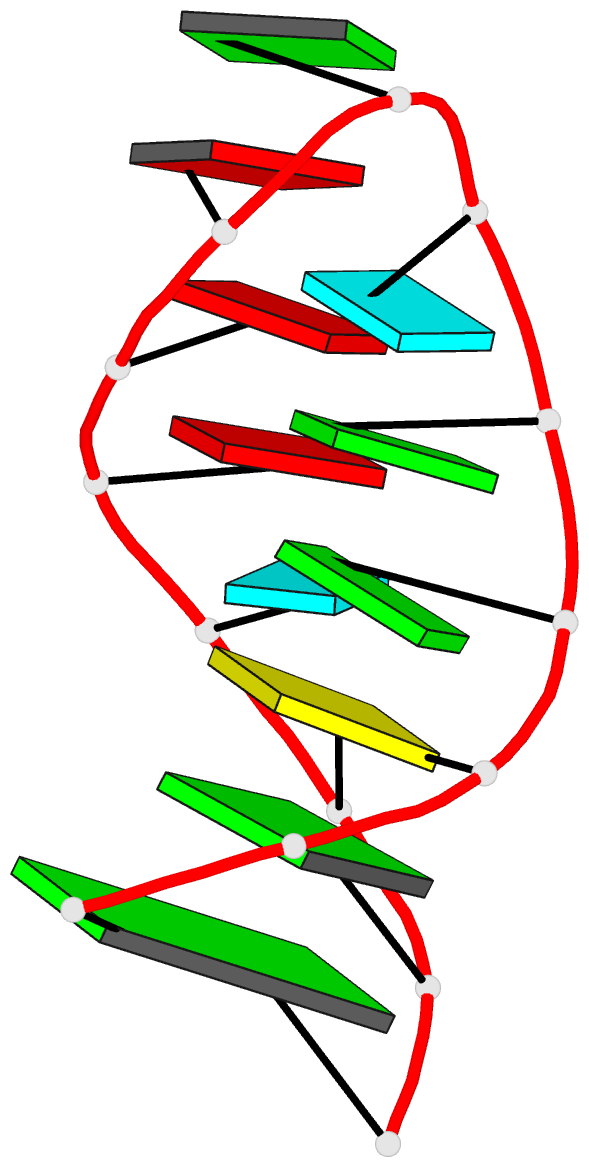
Summary information and primary citation
- PDB-id
- 1fhk; DSSR-derived features in text and JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- NMR structure of the 690 loop of 16 s rrna of e. coli
- Reference
- Morosyuk SV, Cunningham PR, SantaLucia Jr J (2001): "Structure and function of the conserved 690 hairpin in Escherichia coli 16 S ribosomal RNA. II. NMR solution structure." J.Mol.Biol., 307, 197-211. doi: 10.1006/jmbi.2000.4431.
- Abstract
- The solution structure of the conserved 690 hairpin from Escherichia coli 16 S rRNA was determined by NMR spectroscopy. The 690 loop is located at the surface of the 30 S subunit in the platform region and has been implicated in interactions with P-site bound tRNA, E-site mRNA, S11 binding, IF3 binding, and in RNA-RNA interactions with the 790 loop of 16 S rRNA and domain IV of 23 S rRNA. The structure reveals a novel sheared type G690.U697 base-pair with a single hydrogen bond from the G690 amino to U697-04. G691 and A696 also form a sheared pair and U692 forms a U-turn with an H-bond to the A695 non-bridging phosphate oxygen. The sheared pairs and U-turn result in the continuous single-stranded stacking of five residues from 6693 to U697 with their Watson-Crick functional groups exposed in the minor groove. The overall fold of the 690 hairpin is similar to the anticodon loop of tRNA. The structure provides an explanation for chemical protection patterns in the loop upon interaction with tRNA, the 50 S subunit, and S11. In vivo genetic studies demonstrate the functional importance of the motifs observed in the solution structure of the 690 hairpin.