Summary information and primary citation
- PDB-id
- 1d91; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- X-ray (2.1 Å)
- Summary
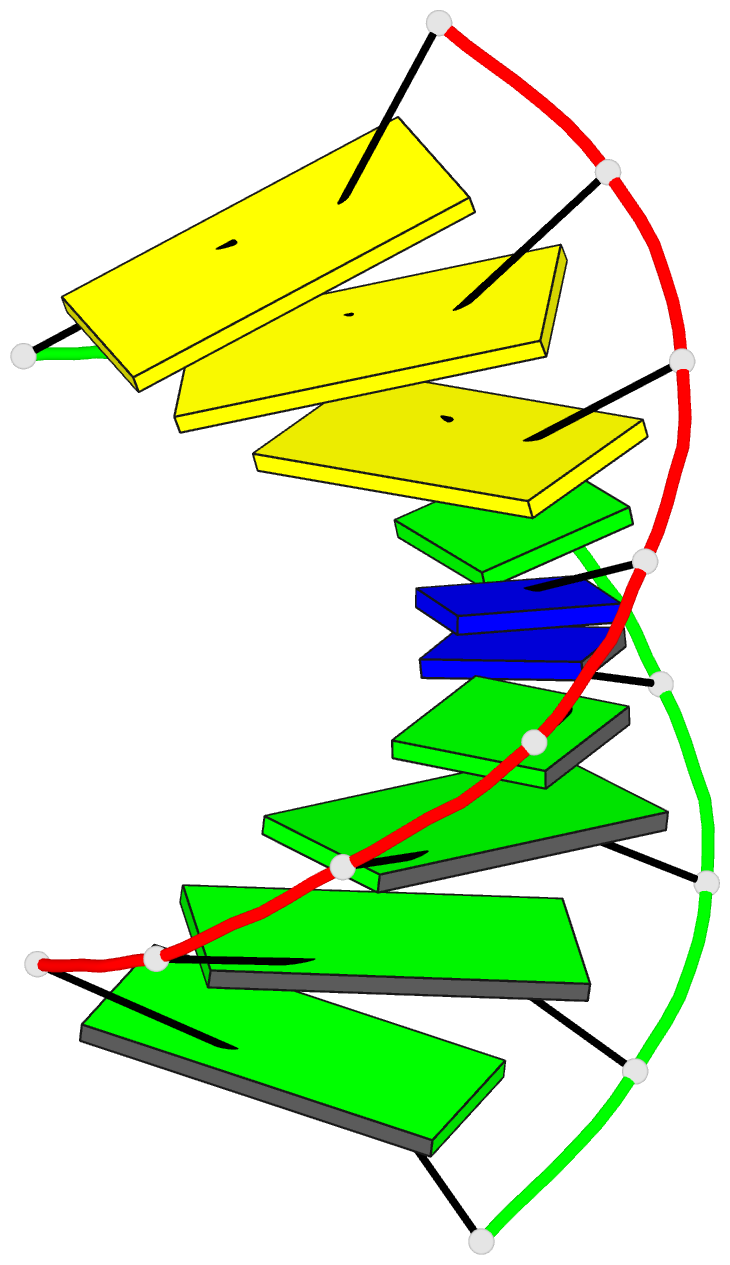
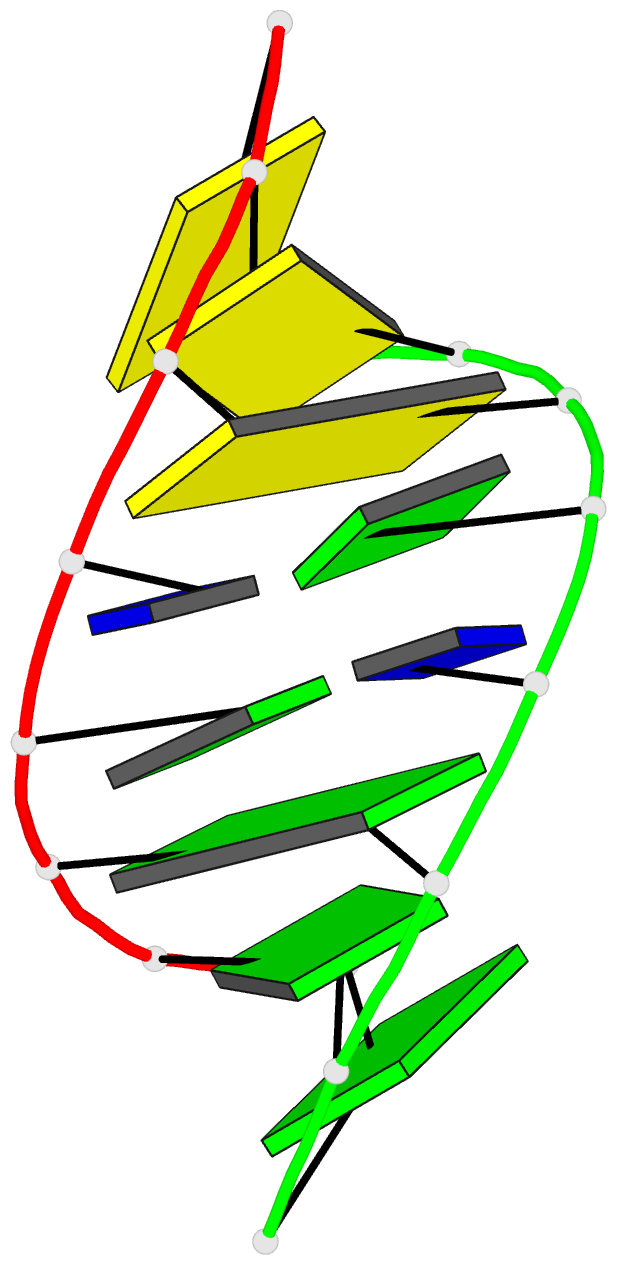
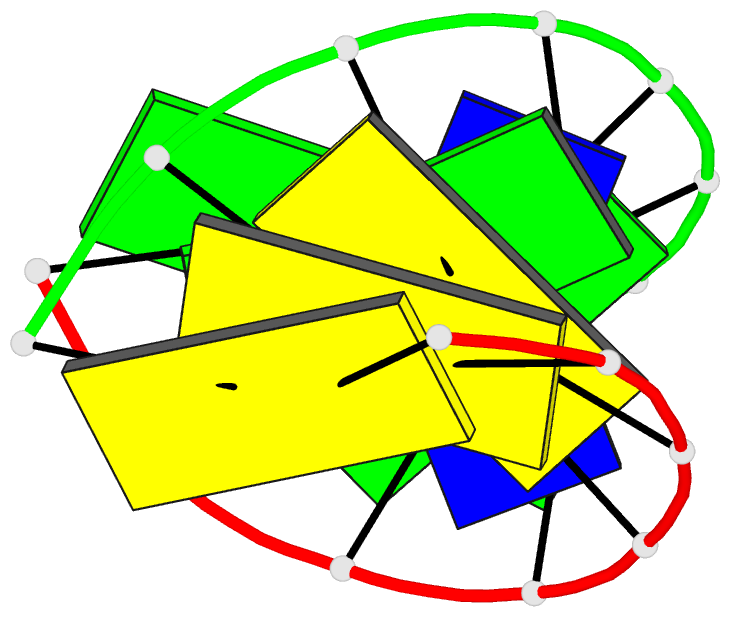
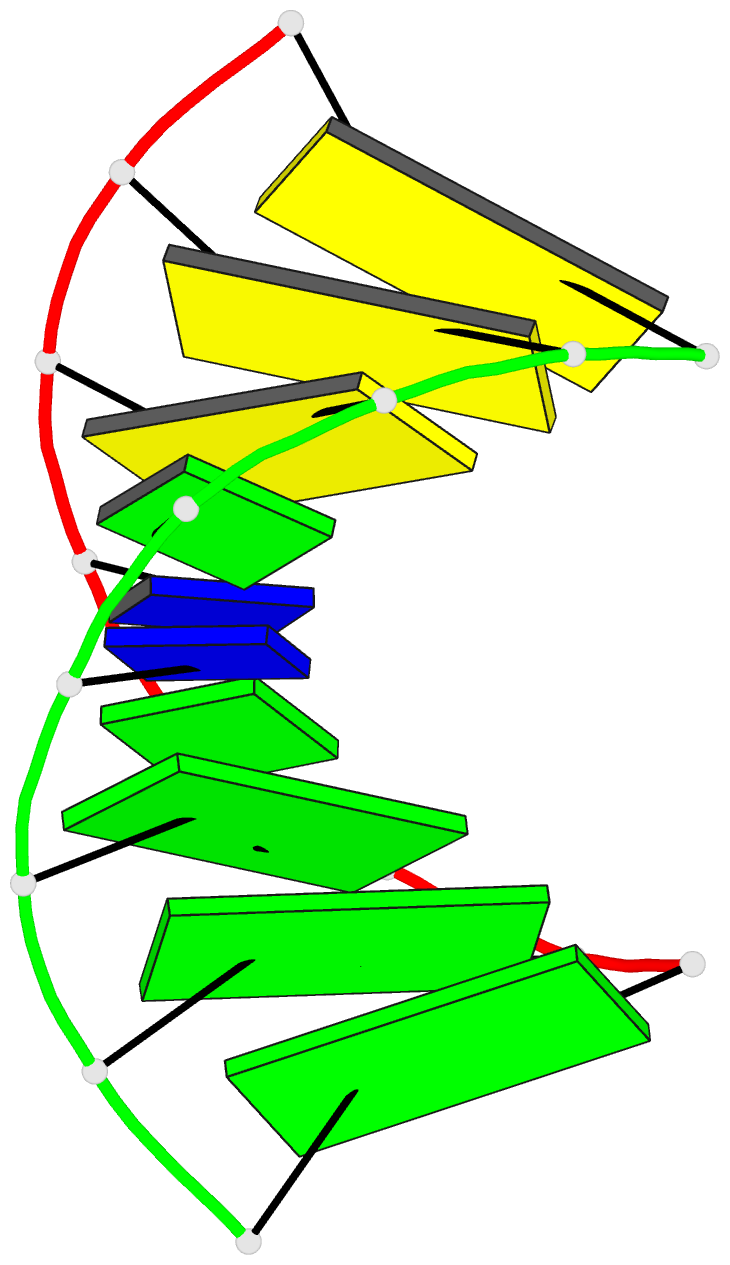
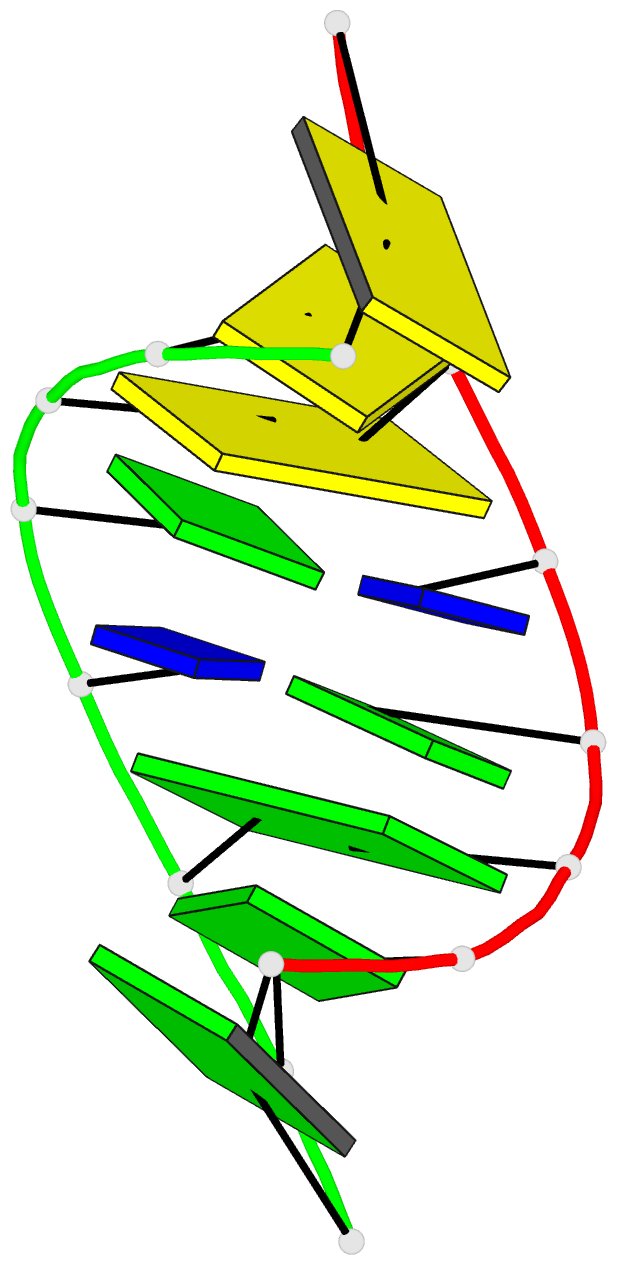
- G.t base pairs in a DNA helix. the crystal structure of d(g-g-g-g-t-c-c-c)
- Reference
- Kneale G, Brown T, Kennard O, Rabinovich D (1985): "G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C)." J.Mol.Biol., 186, 805-814. doi: 10.1016/0022-2836(85)90398-5.
- Abstract
- The synthetic deoxyoctanucleotide d(G-G-G-G-T-C-C-C) crystallizes as an A-type DNA double helix containing two adjacent G . T base-pair mismatches. The structure has been refined to an R-factor of 14% at 2.1 A resolution with 104 solvent molecules located. The two G . T mismatches adopt the "wobble" form of base-pairing. The mismatched bases are linked by a network of water molecules interacting with the exposed functional groups in both the major and minor grooves. The presence of two mispaired bases in the octamer has surprisingly little effect on the global structure of the helix or the backbone and glycosidic torsional angles. Base stacking around the mismatch is perturbed, but the central G-T step shows particularly good base overlap, which may contribute to the relatively high stability of this oligomer.