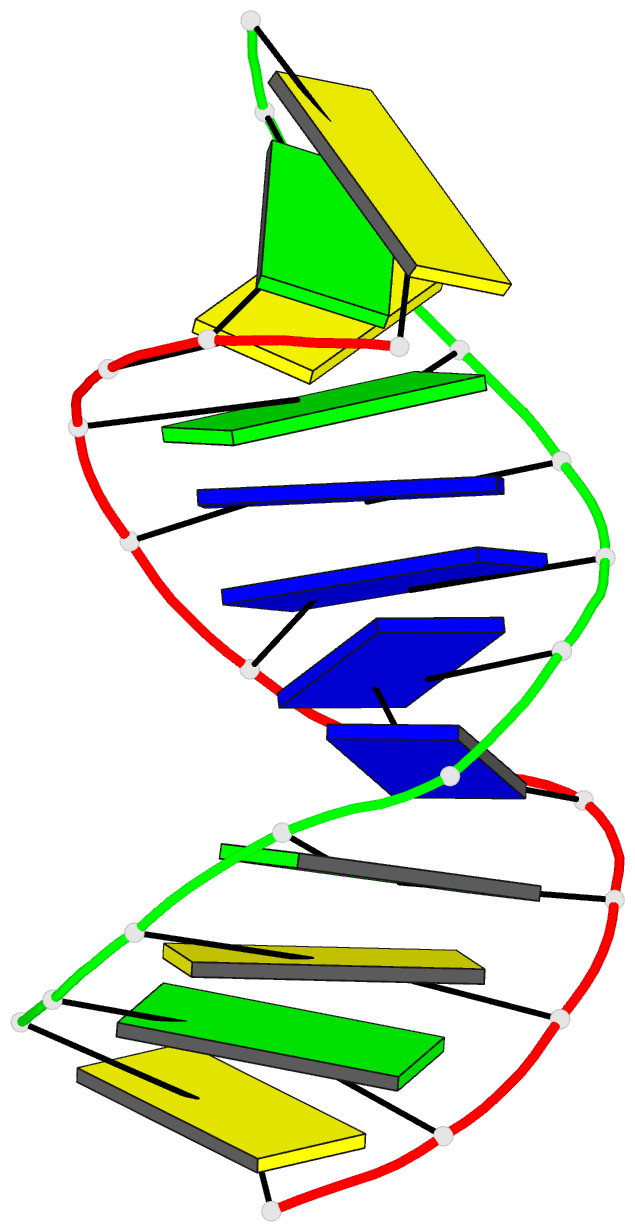
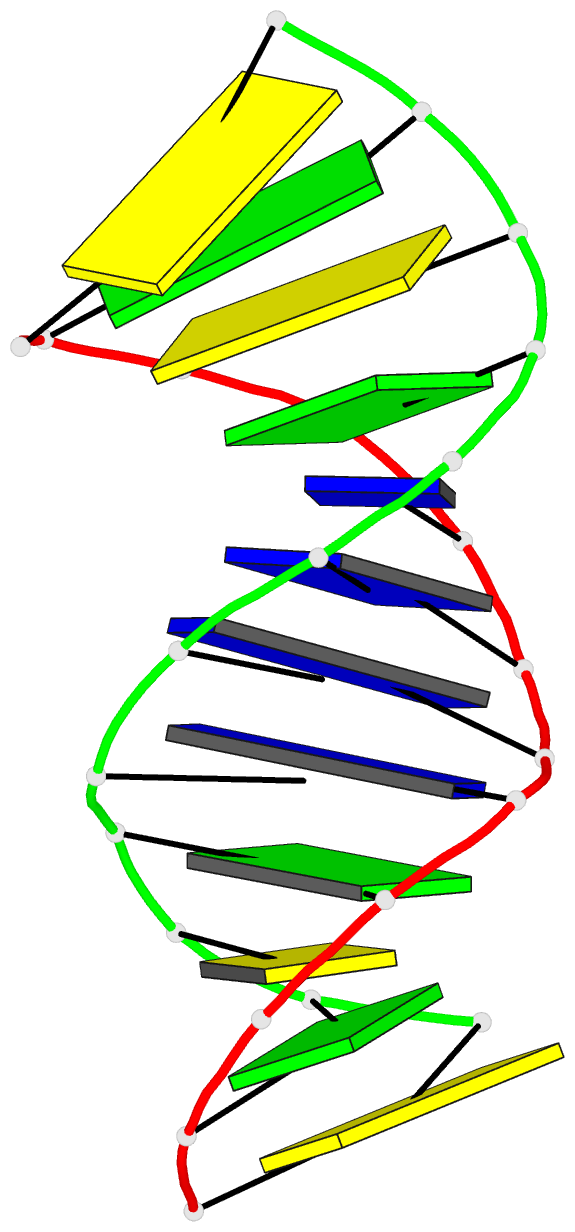
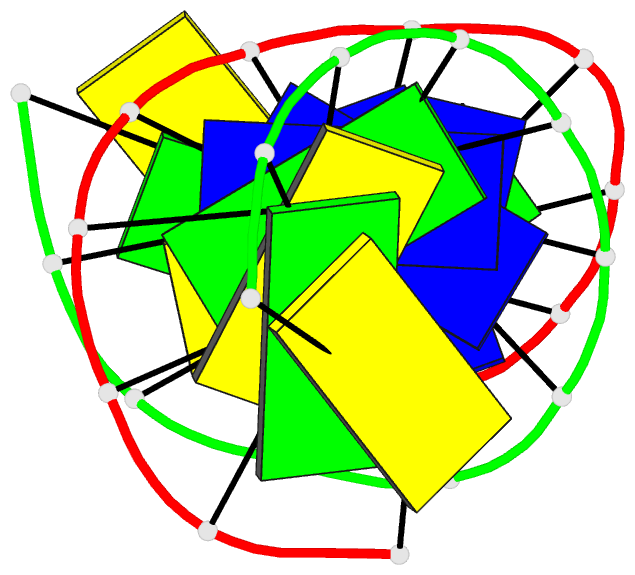
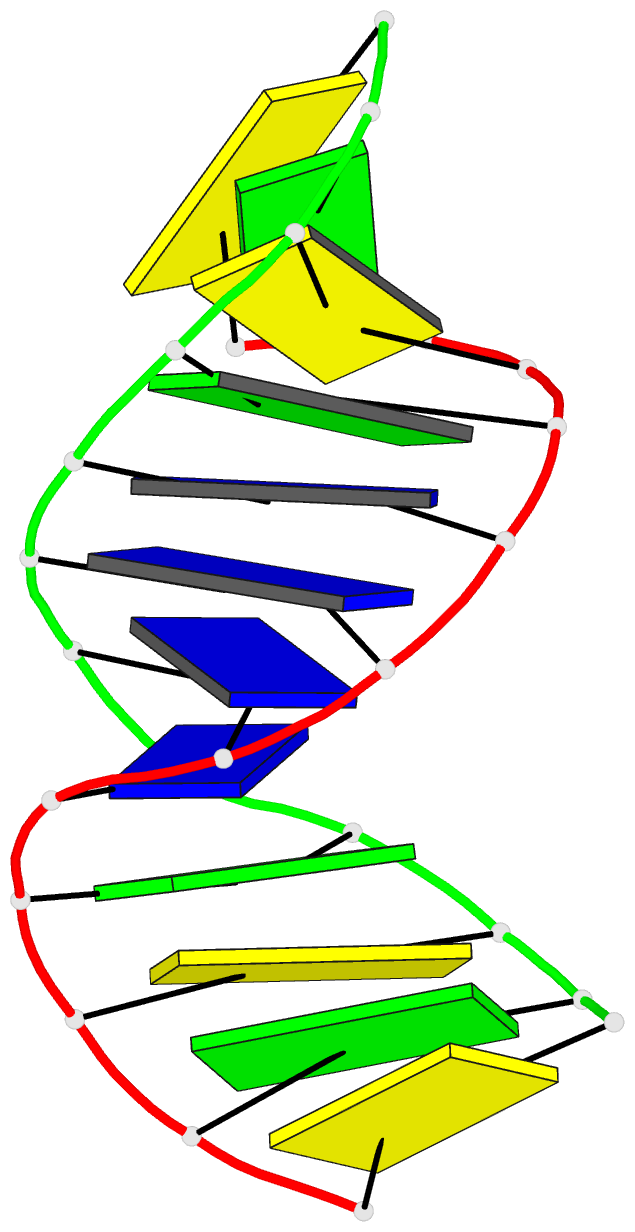
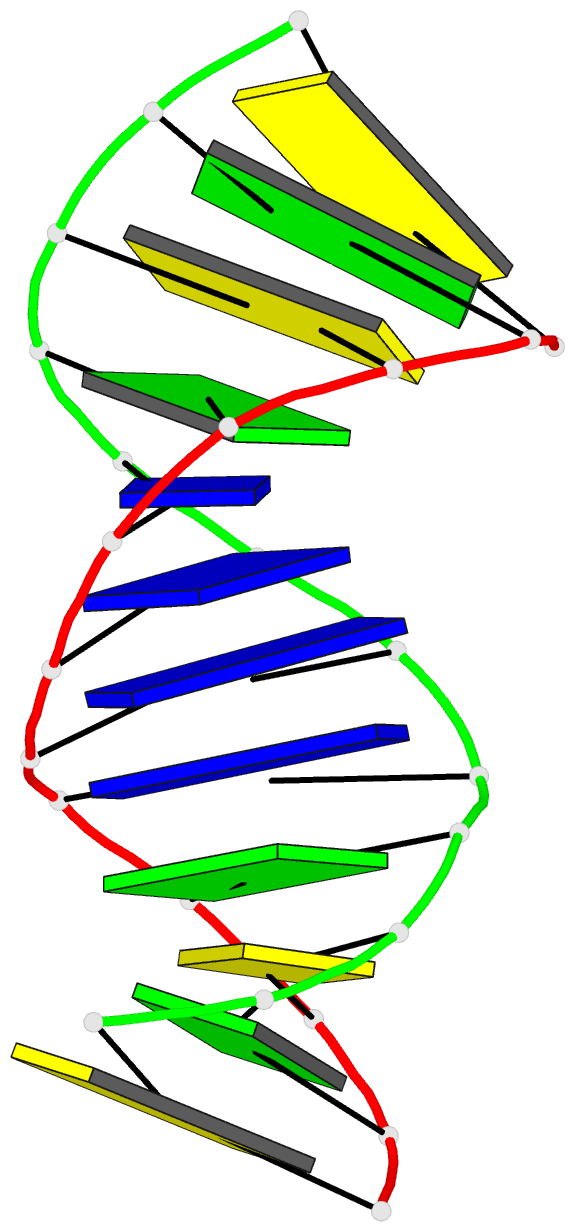
Summary information and primary citation
- PDB-id
- 1cx5; DSSR-derived features in text and JSON formats
- Class
- DNA-RNA hybrid
- Method
- NMR
- Summary
- Antisense DNA-RNA hybrid containing modified backbone
- Reference
- Yang X, Han X, Cross C, Bare S, Sanghvi Y, Gao X (1999): "NMR structure of an antisense DNA.RNA hybrid duplex containing a 3'-CH(2)N(CH(3))-O-5' or an MMI backbone linker." Biochemistry, 38, 12586-12596. doi: 10.1021/bi990456x.
- Abstract
- The solution structure of an antisense DNA.RNA hybrid duplex, d(CGCGTT-MMI-TTGCGC).r(GCGCAAAACGCG) (designated R4), containing an MMI backbone linker [3'-CH(2)N(CH(3))-O5'], is elucidated. The structural details of the MMI linker, its structural effects on the neighboring residues, and the molecular basis of the MMI effects are examined. The lipophilic N-methyl group of MMI is peripheral to the helix, assuming a conformation that is most stable with regard to the N-O torsion angle. The MMI linker promotes a 3'-endo conformation for the sugar moieties at both 3'- and 5'-adjacent positions and a backbone kink involving distant residues along the 3'-direction. Comparison of R4 with other analogous hybrid duplexes previously studied in this laboratory reveals a new family of low-energy helical conformations that can be accommodated in stable duplexes and a common feature of C3'-modified sugars for adopting a C3'-endo pucker. The results of these studies emphasize the interplay of several factors that govern the formation of stable hybrid duplexes and provide a basis for the understanding of the biological role of the MMI modifications, which are important building blocks for a family of promising chimeric antisense oligonucleotides.