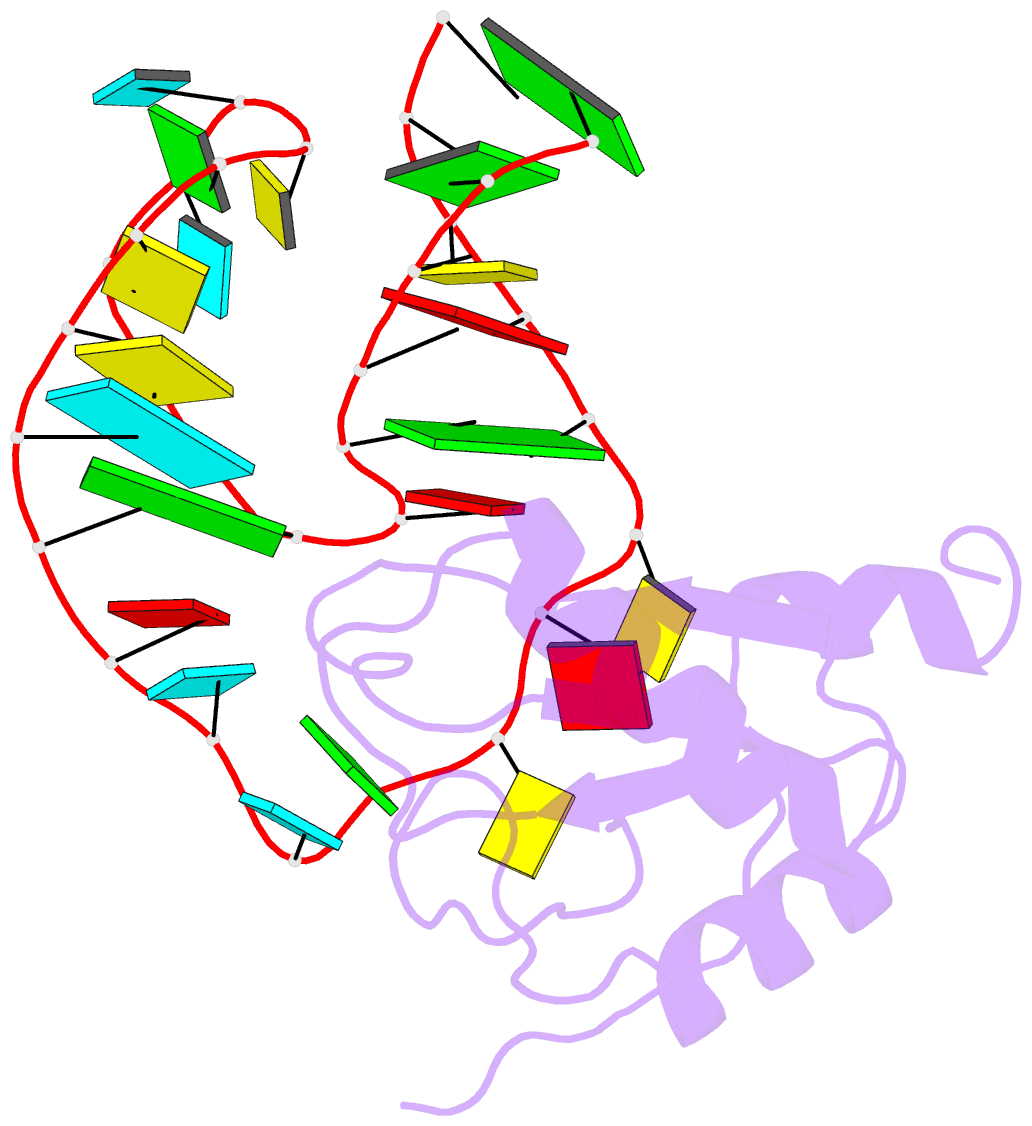
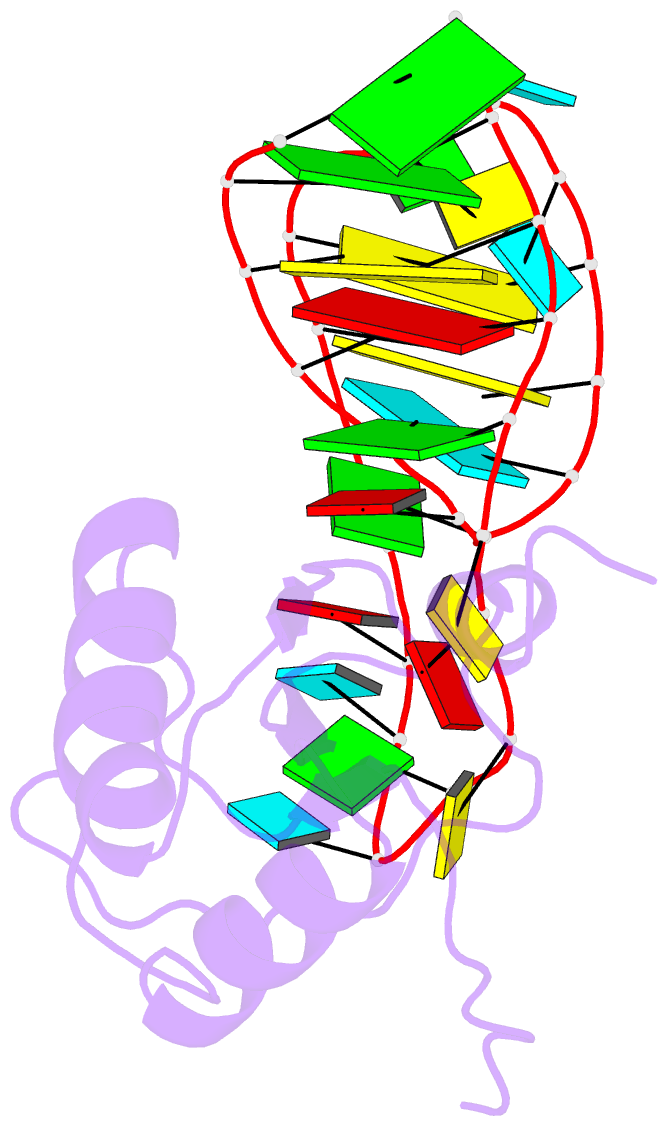
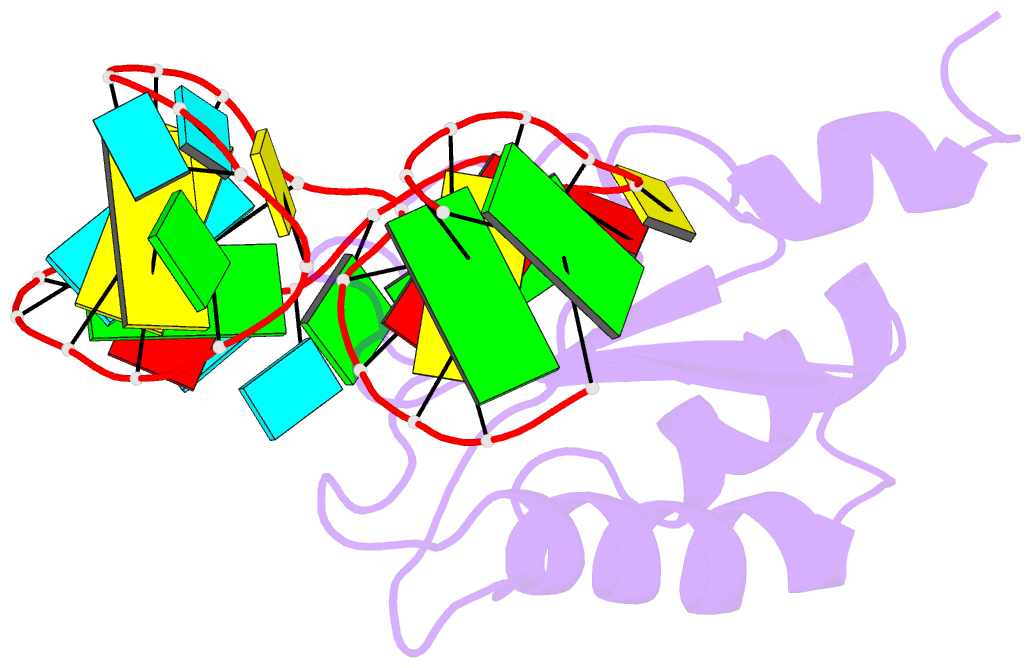
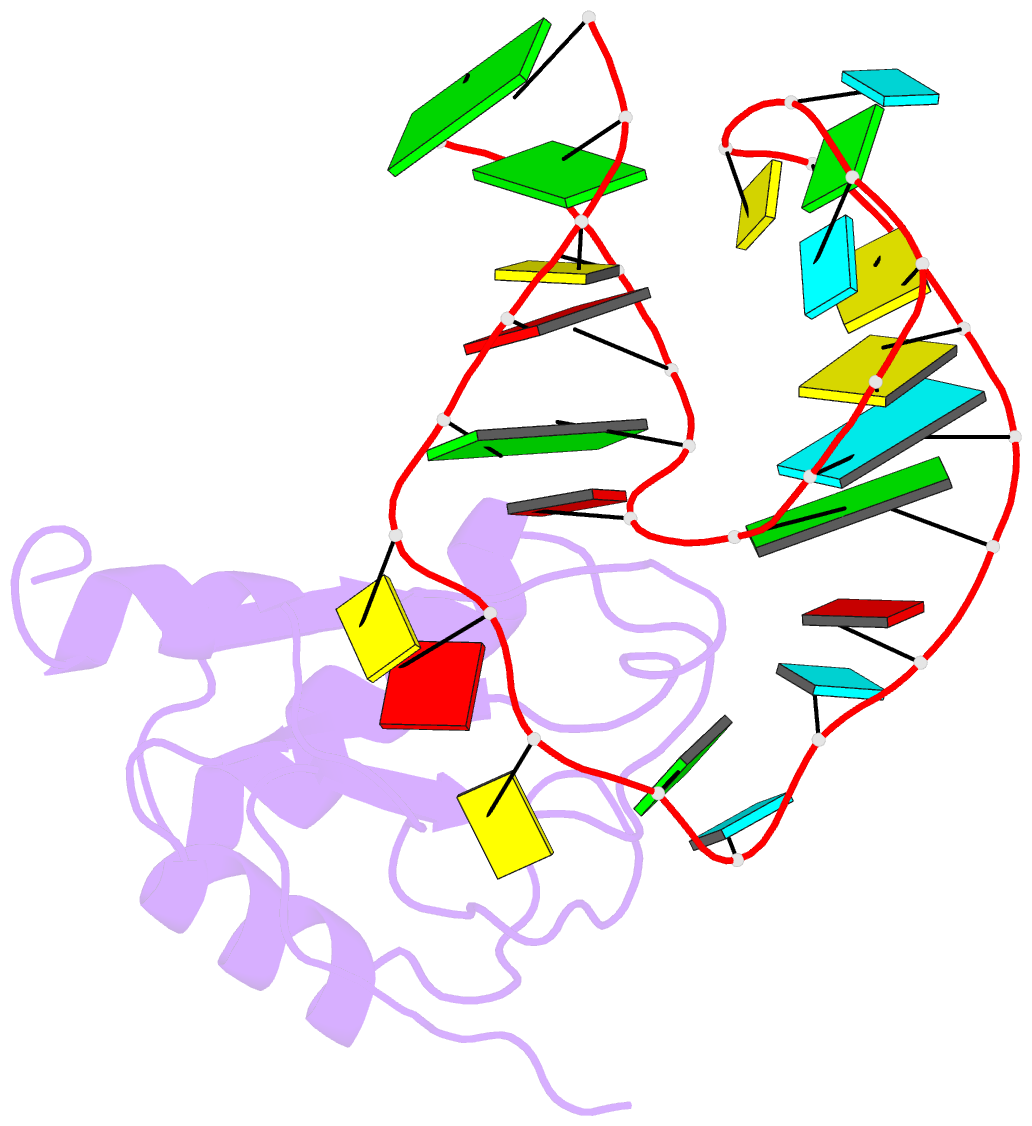
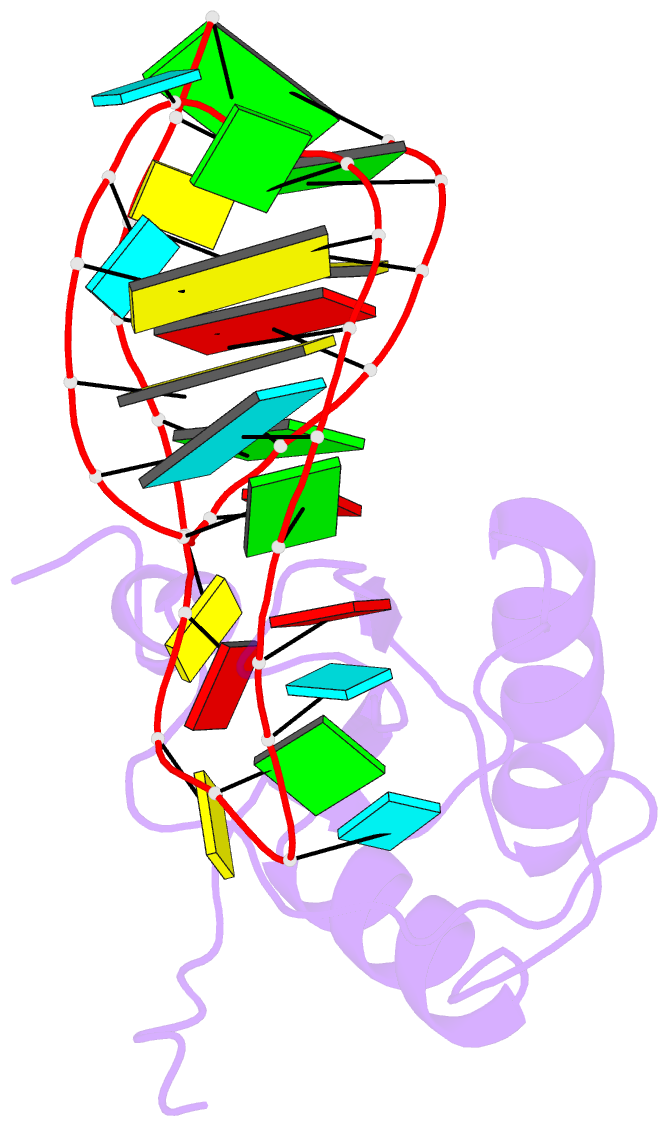
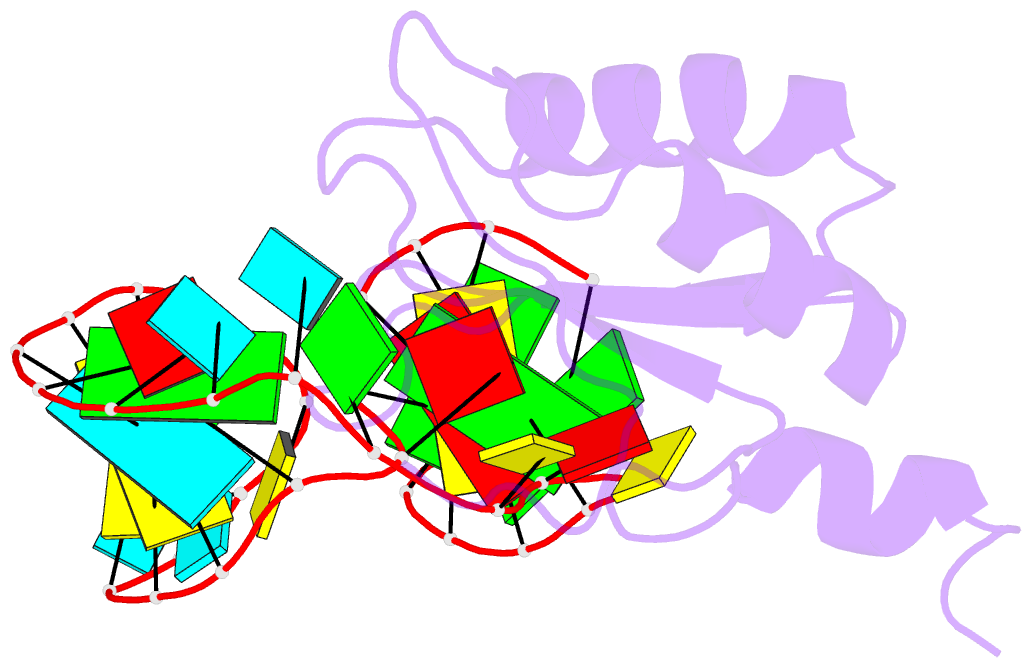
Summary information and primary citation
- PDB-id
- 1aud; DSSR-derived features in text and JSON formats
- Class
- RNA binding protein-RNA
- Method
- NMR
- Summary
- U1a-utrrna, NMR, 31 structures
- Reference
- Allain FH, Howe PW, Neuhaus D, Varani G (1997): "Structural basis of the RNA-binding specificity of human U1A protein." EMBO J., 16, 5764-5774. doi: 10.1093/emboj/16.18.5764.
- Abstract
- The RNP domain is a very common eukaryotic protein domain involved in recognition of a wide range of RNA structures and sequences. Two structures of human U1A in complex with distinct RNA substrates have revealed important aspects of RNP-RNA recognition, but have also raised intriguing questions concerning the origin of binding specificity. The beta-sheet of the domain provides an extensive RNA-binding platform for packing aromatic RNA bases and hydrophobic protein side chains. However, many interactions between functional groups on the single-stranded nucleotides and residues on the beta-sheet surface are potentially common to RNP proteins with diverse specificity and therefore make only limited contribution to molecular discrimination. The refined structure of the U1A complex with the RNA polyadenylation inhibition element reported here clarifies the role of the RNP domain principal specificity determinants (the variable loops) in molecular recognition. The most variable region of RNP proteins, loop 3, plays a crucial role in defining the global geometry of the intermolecular interface. Electrostatic interactions with the RNA phosphodiester backbone involve protein side chains that are unique to U1A and are likely to be important for discrimination. This analysis provides a novel picture of RNA-protein recognition, much closer to our current understanding of protein-protein recognition than that of DNA-protein recognition.