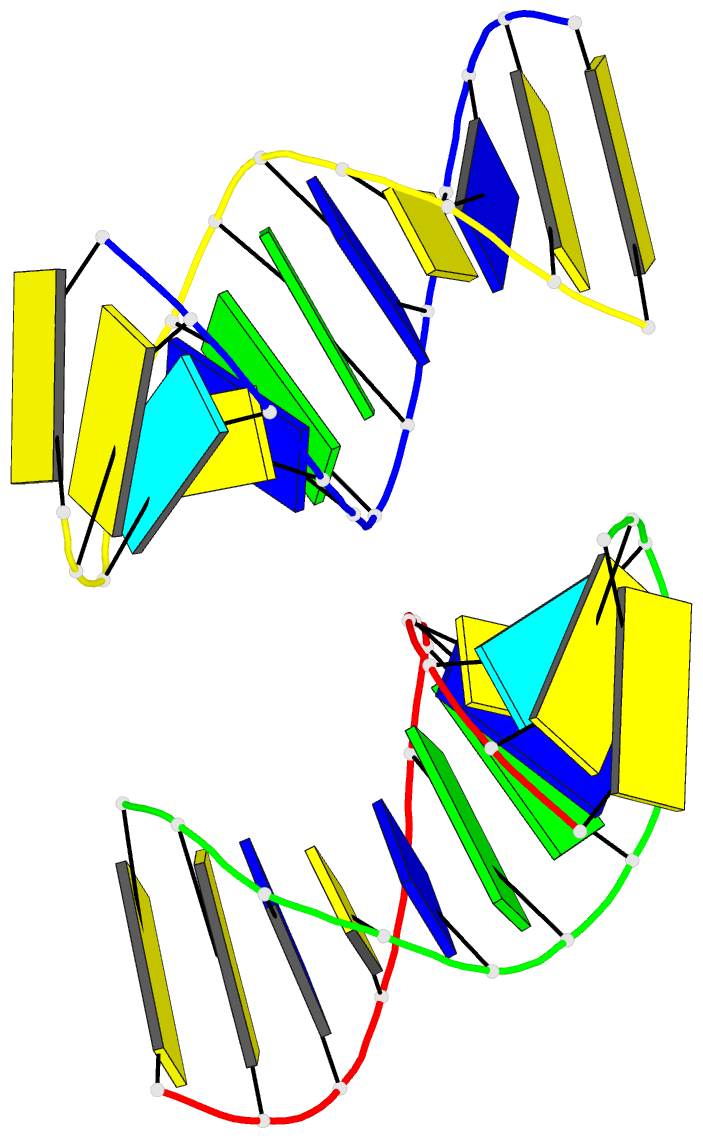
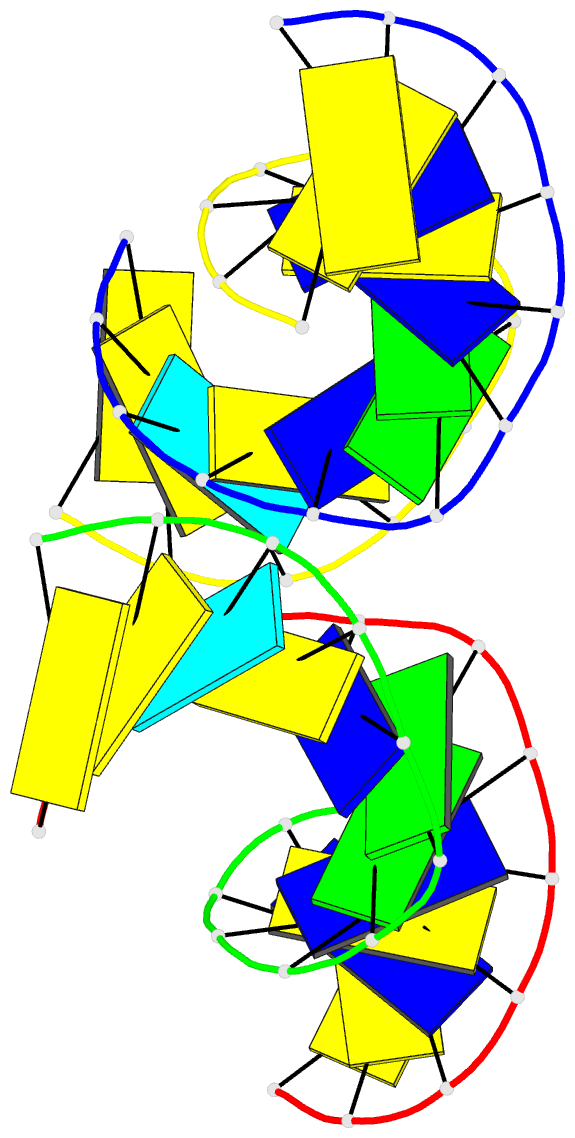
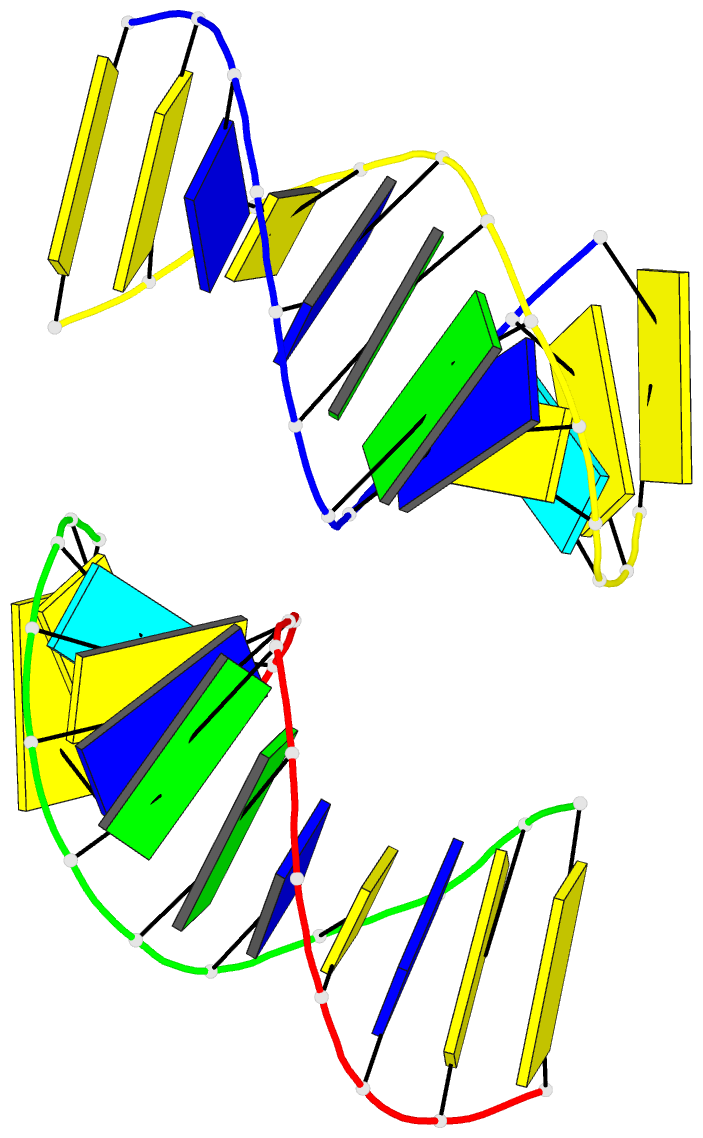
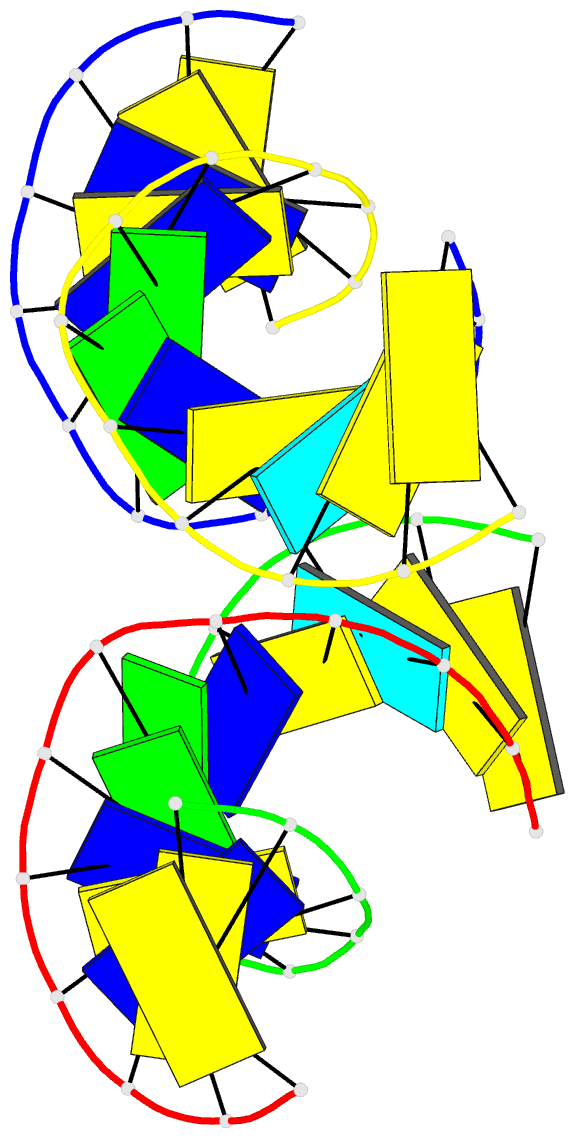
Summary information and primary citation
- PDB-id
- 1aio; DSSR-derived features in text and JSON formats
- Class
- DNA
- Method
- X-ray (2.6 Å)
- Summary
- Crystal structure of a double-stranded DNA containing the major adduct of the anticancer drug cisplatin
- Reference
- Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ (1995): "Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin." Nature, 377, 649-652. doi: 10.1038/377649a0.
- Abstract
- The success of cisplatin in cancer chemotherapy derives from its ability to crosslink DNA and alter the structure. Most cisplatin-DNA adducts are intrastrand d(GpG) and d(ApG) crosslinks, which unwind and bend the duplex to facilitate the binding of proteins that contain one or more high-mobility group (HMG) domains. When HMG-domain proteins such as HMG1, IXR (intrastrand-crosslink recognition) protein from yeast, or human upstream-binding factor (hUBF) bind cisplatin intrastrand crosslinks, they can be diverted from their natural binding sites on the genome and shield the adducts from excision repair. These activities sensitize cells to cisplatin and contribute to its cytotoxic properties. Crystallographic information about the structure of cisplatin-DNA adducts has been limited to short single-stranded deoxyoligonucleotides such as cis-[Pt(NH3)2(d(pGpG))]. Here we describe the X-ray structure at 2.6 A resolution of a double-stranded DNA dodecamer containing this adduct. Our information provides, to our knowledge, the first crystallographic look at a platinated DNA duplex and should help the design of new platinum and other metal crosslinking antitumour drug candidates. Moreover, the structure reveals a unique fusion of A- and B-type DNA segments that could be of more general importance.