Summary information and primary citation
- PDB-id
-
1a34;
SNAP-derived features in text and
JSON formats
- Class
- virus-RNA
- Method
- X-ray (1.81 Å)
- Summary
- Satellite tobacco mosaic virus-RNA complex
- Reference
-
Larson SB, Day J, Greenwood A, McPherson A (1998):
"Refined
structure of satellite tobacco mosaic virus at 1.8 A
resolution." J.Mol.Biol.,
277, 37-59. doi: 10.1006/jmbi.1997.1570.
- Abstract
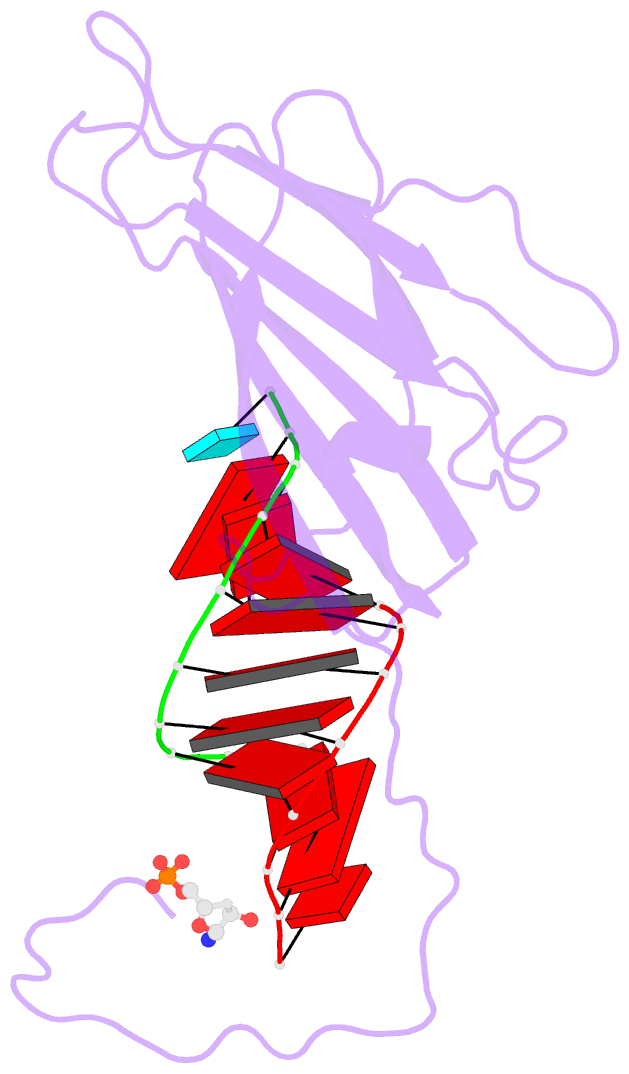
- The molecular structure of satellite tobacco mosaic
virus (STMV) has been refined to 1.8 A resolution using
X-ray diffraction data collected from crystals grown in
microgravity. The final R value was 0.179 and Rfree was
0.184 for 219,086 independent reflections. The final model
of the asymmetric unit contained amino acid residues 13 to
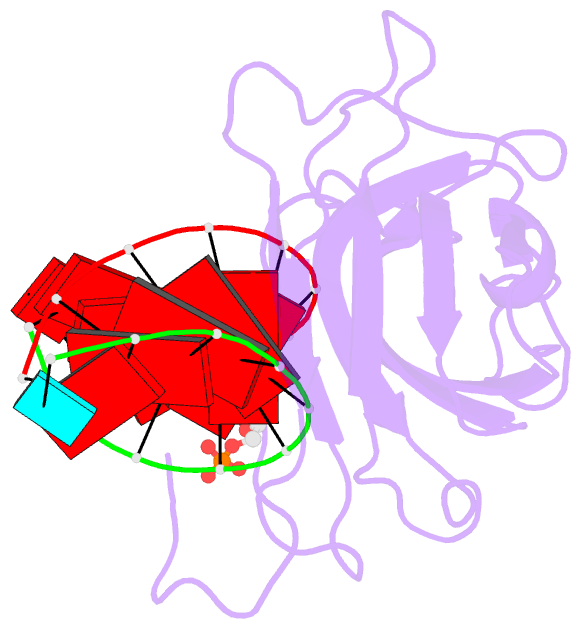
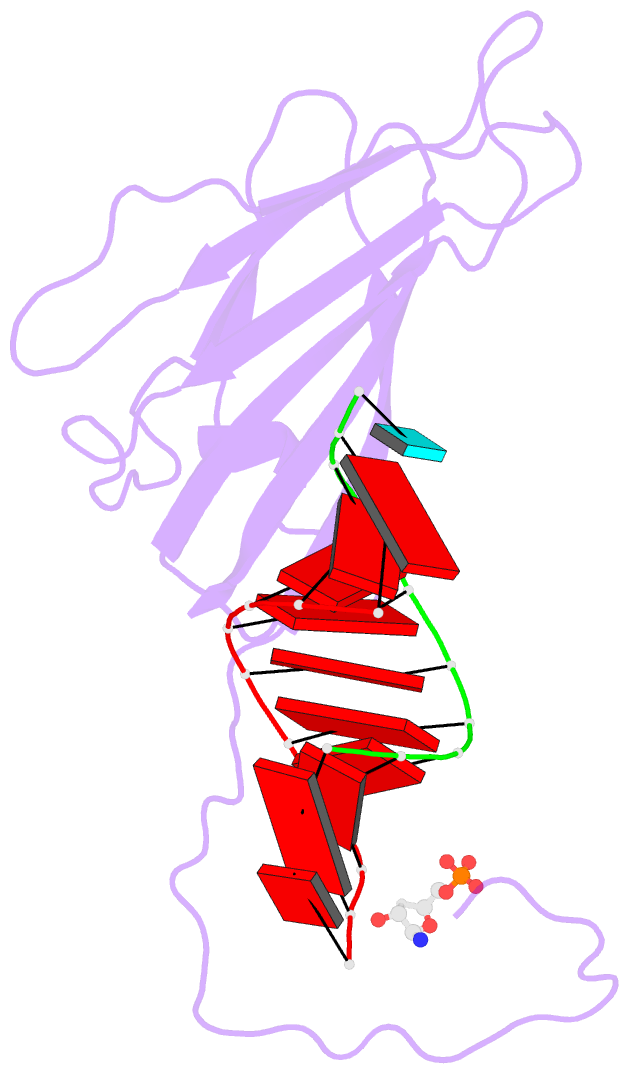
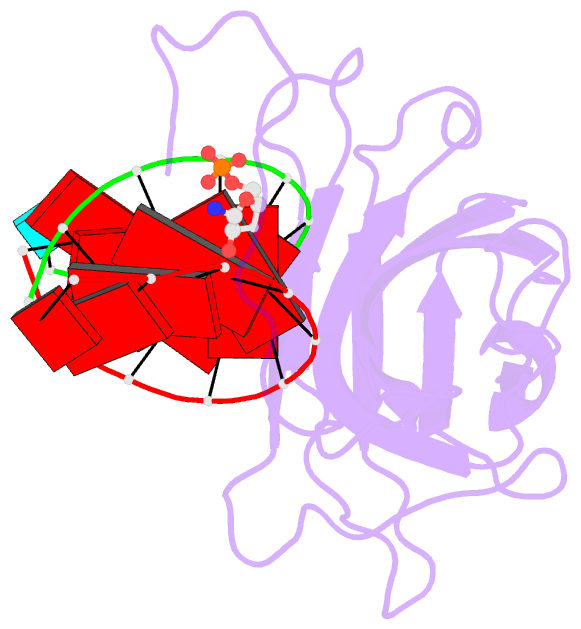
159 of a coat protein monomer, 21 nucleotides, a sulfate
ion, and 168 water molecules. The nucleotides were
visualized as 30 helical segments of nine base-pairs with
an additional base stacked at each 3' end, plus a "free"
nucleotide, not belonging to the helical segments, but
firmly bound by the protein. Sulfate ions are located
exactly on 5-fold axes and each is coordinated by ten
asparagine side-chains. Of the 10,080 structural waters,
168 per asymmetric unit, about 20% serve to bridge the
macromolecular components at protein-protein and
protein-nucleic acid interfaces. Binding of RNA to the
protein involves some salt linkages, particularly to the
phosphate of the free nucleotide, but the major
contribution is from an intricate network of hydrogen
bonds. There are numerous water molecules in the
RNA-protein interface, many serving as intermediate
hydrogen bond bridges. The sugar-phosphate backbone
contributes most of the donors and acceptors for the RNA.
The helical RNA conformation is nearest that of A form DNA.
The central region of a helical segment is most extensively
involved in contacts with protein, and exhibits low thermal
parameters which increase dramatically toward the ends. The
visible RNA represents approximately 59% of the total
nucleic acid in the virion and is derived from the
single-stranded genome, which has folded upon itself to
form helical segments. Linking of the helices and the free
nucleotides in a contiguous and efficient manner severely
restricts the disposition of the remaining, unseen nucleic
acid. Using the remaining nucleotides it is possible to
fold the RNA according to motifs that provide a periodic
distribution of RNA structural elements compatible with the
icosahedrally symmetrical arrangement seen in the
crystallographic structure. The intimate relationship
between protein and nucleic acid in STMV suggests an
assembly pathway based on the cooperative and coordinated
co-condensation of RNA with capsid protein dimers.