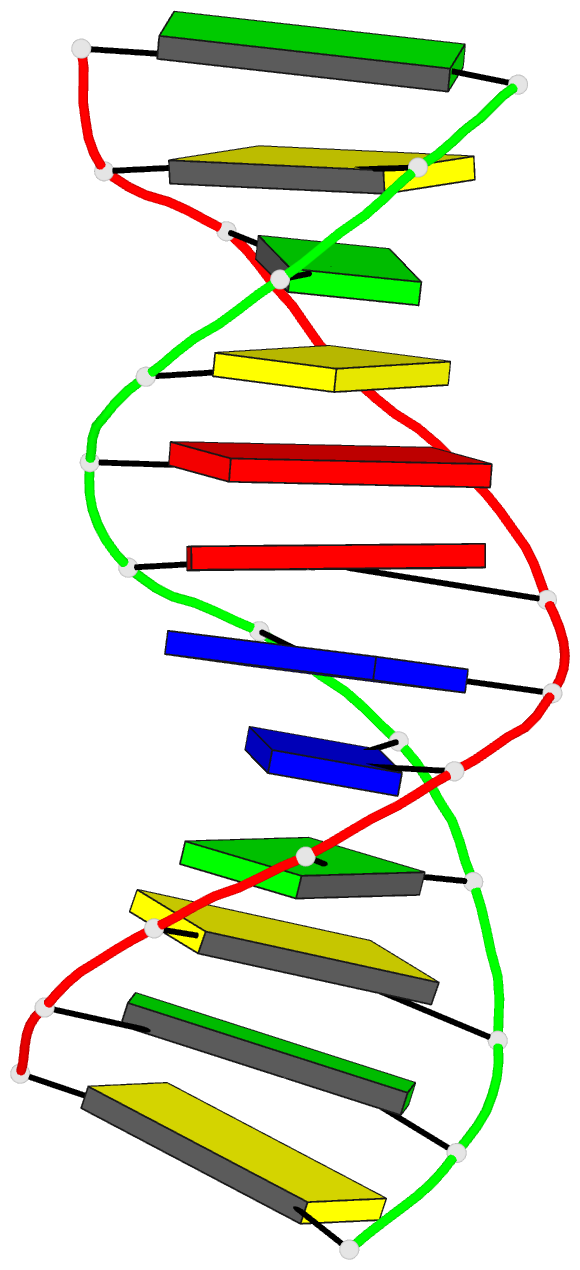
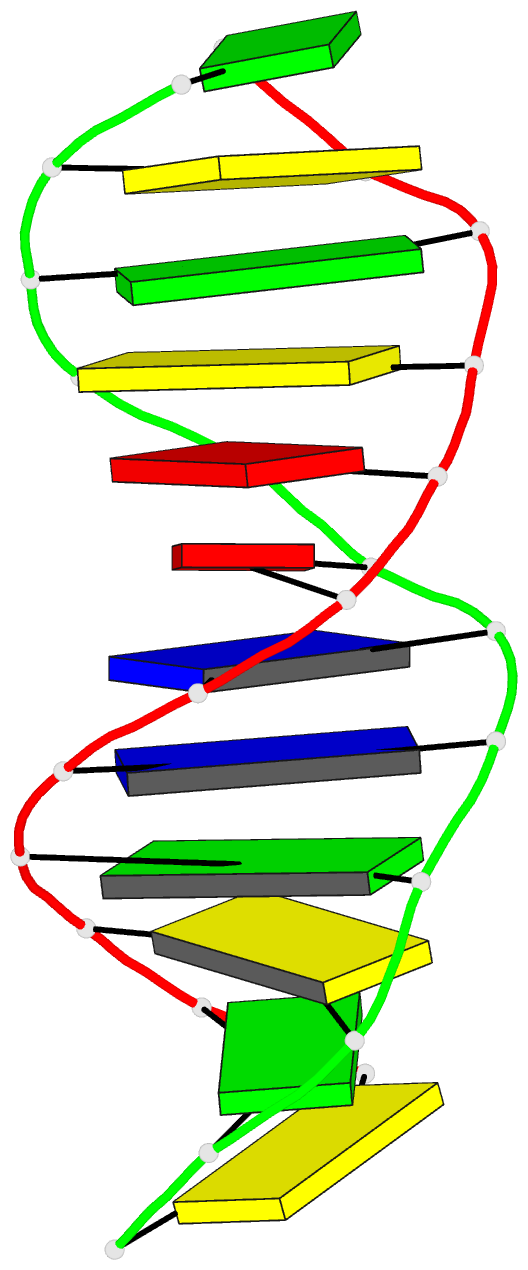
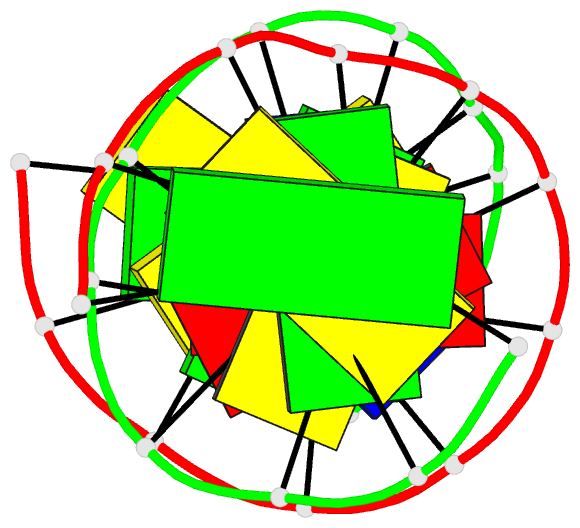
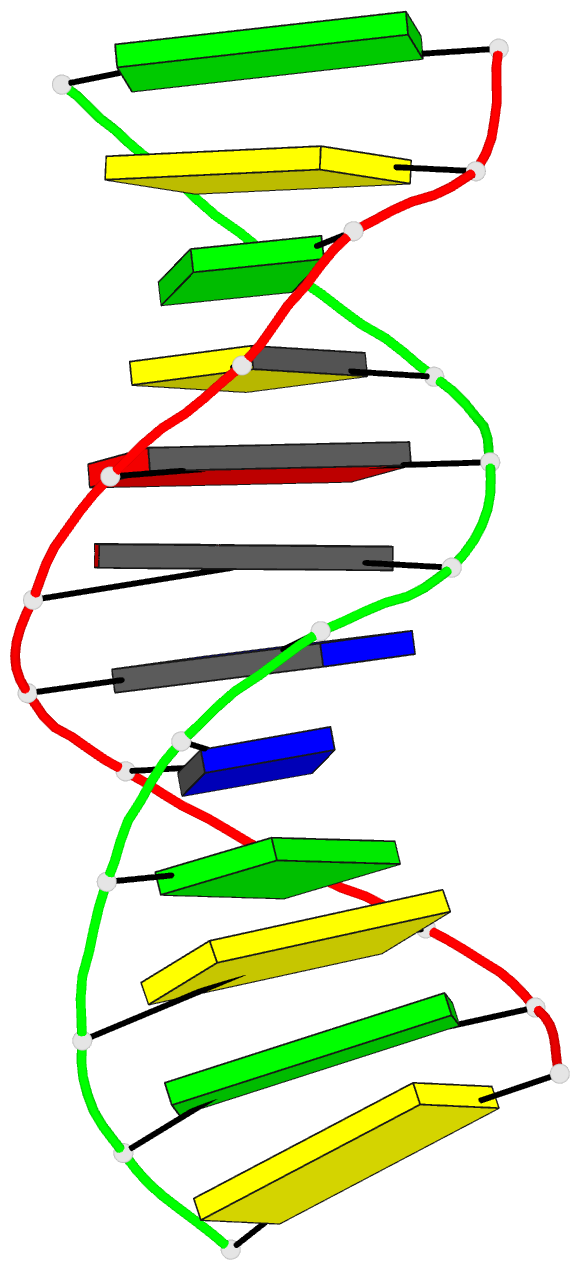
Summary information and primary citation
- PDB-id
-
194d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- X-ray (2.3 Å)
- Summary
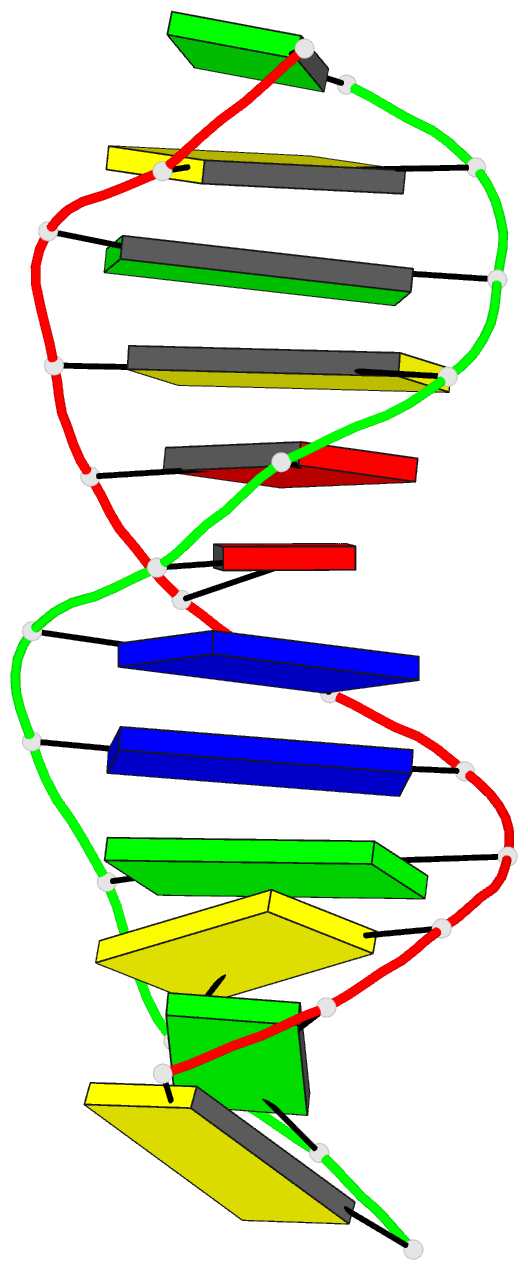
- X-ray structures of the b-DNA dodecamer d(cgcgttaacgcg)
with an inverted central tetranucleotide and its netropsin
complex
- Reference
-
Balendiran K, Rao ST, Sekharudu CY, Zon G, Sundaralingam
M (1995): "X-ray
structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an
inverted central tetranucleotide and its netropsin
complex." Acta Crystallogr.,Sect.D,
51, 190-198. doi: 10.1107/S0907444994010759.
- Abstract
- The crystal structures of the B-DNA dodecamer
d(CGCGTTAACGCG) duplex (T2A2), with the inverted
tetranucleotide core from the duplex d(CGCGAATTCGCG) [A2T2,
Dickerson & Drew (1981). J. Mol. Biol. 149, 761-768], and
its netropsin complex (T2A2-N) have been determined at 2.3
A resolution. The crystals are orthorhombic, space group
P2(1)2(1)2(1), unit-cell dimensions of a = 25.7, b = 40.5
and c = 67.0 A, for T2A2 and a = 25.49, b = 40.87, c =
67.02 A for T2A2-N and are isomorphous with A2T2. The
native T2A2 structure, with 70 water molecules had a final
R value of 0.15 for 1522 reflections (F > 2sigma), while
for the netropsin complex, with 87 water molecules, the R
value was 0.16 for 2420 reflections. In T2A2, a
discontinuous string of zig-zagging water molecules hydrate
the narrow A.T minor groove. In T2A2-N, netropsin binds in
one orientation in the minor groove, covering the TTAA
central region, by displacing the string of waters, forming
the majority of hydrogen bonds with DNA atoms in one
strand, and causing very little perturbation of the native
structure. The helical twist angle in T2A2 is largest at
the duplex center, corresponding to the cleavage site by
the restriction enzymes HpaI and HincII. The sequence
inversion AATT-->TTAA of the tetranucleotide at the
center of the molecule results in a different path for the
local helix axis in T2A2 and A2T2 but the overall bending
is similar in both cases.