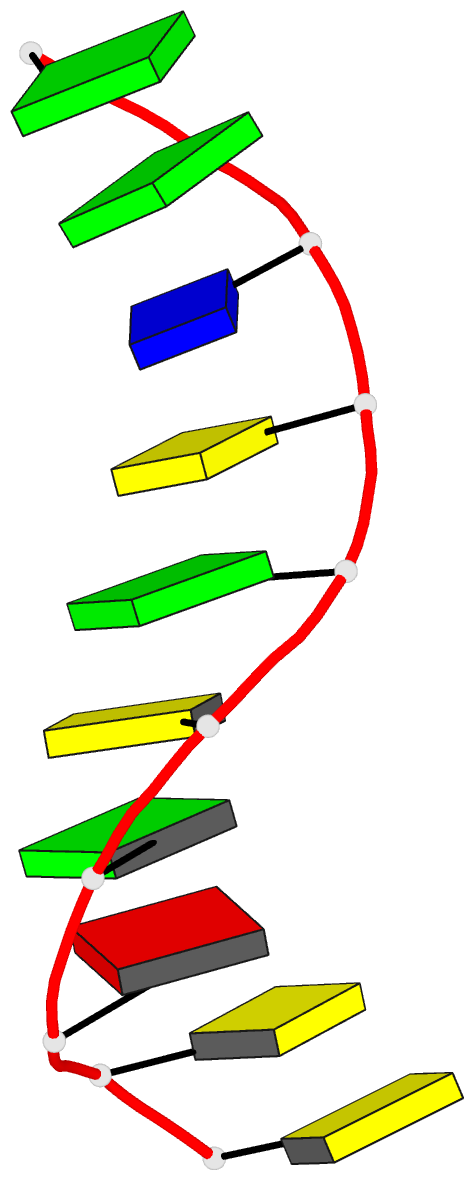
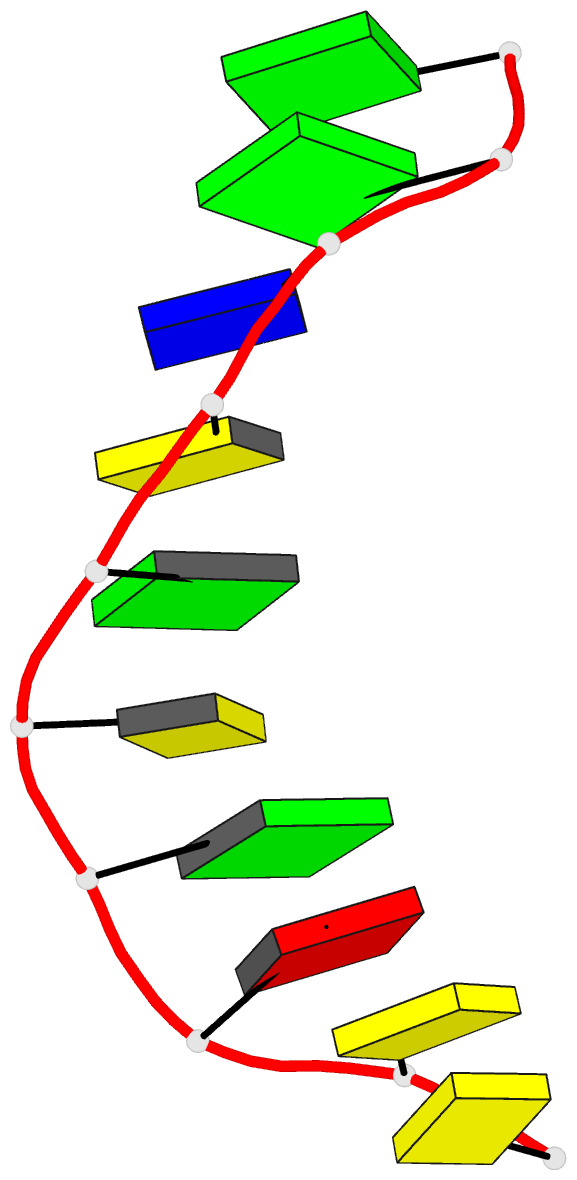
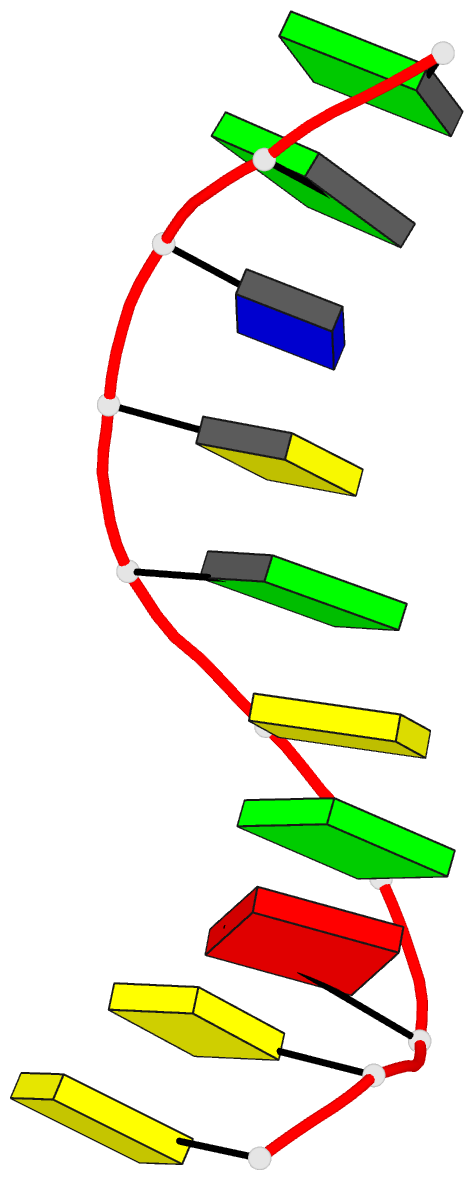
Summary information and primary citation
- PDB-id
-
183d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- X-ray (1.6 Å)
- Summary
- X-ray structure of a DNA decamer containing 7,
8-dihydro-8-oxoguanine
- Reference
-
Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller
EM, Rich A, Essigmann JM, Williams LD (1995): "X-ray
structure of a DNA decamer containing
7,8-dihydro-8-oxoguanine."
Proc.Natl.Acad.Sci.USA, 92,
719-723. doi: 10.1073/pnas.92.3.719.
- Abstract
- We have determined the x-ray structure of a DNA
fragment containing 7,8-dihydro-8-oxoguanine (G(O)). The
structure of the duplex form of d(CCAGOCGCTGG) has been
determined to 1.6-A resolution. The results demonstrate
that GO forms Watson-Crick base pairs with the opposite C
and that G(O) is in the anti conformation. Structural
perturbations induced by C.G(O)anti base pairs are subtle.
The structure allows us to identify probable elements by
which the DNA repair protein MutM recognizes its
substrates. Hydrogen bond donors/acceptors within the major
groove are the most likely element. In that groove, the
pattern of hydrogen-bond donors/acceptors of C.G(O)anti is
unique. Additional structural analysis indicates that
conversion of G to G(O) would not significantly influence
the glycosidic torsion preference of the nucleoside. There
is no steric interaction of the 8-oxygen of G(O) with the
phospho-deoxyribose backbone.