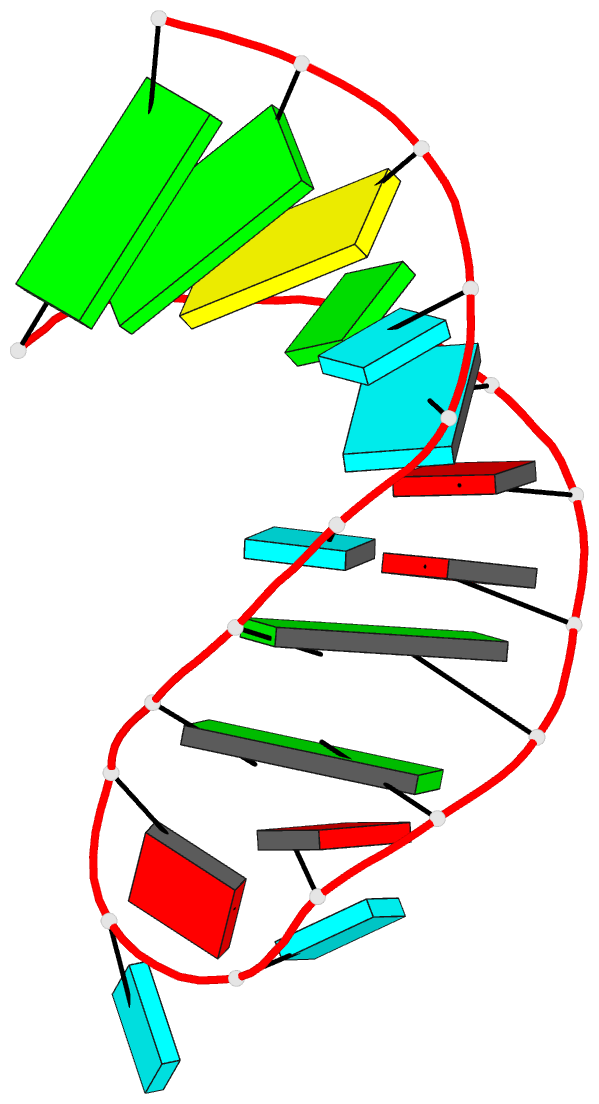
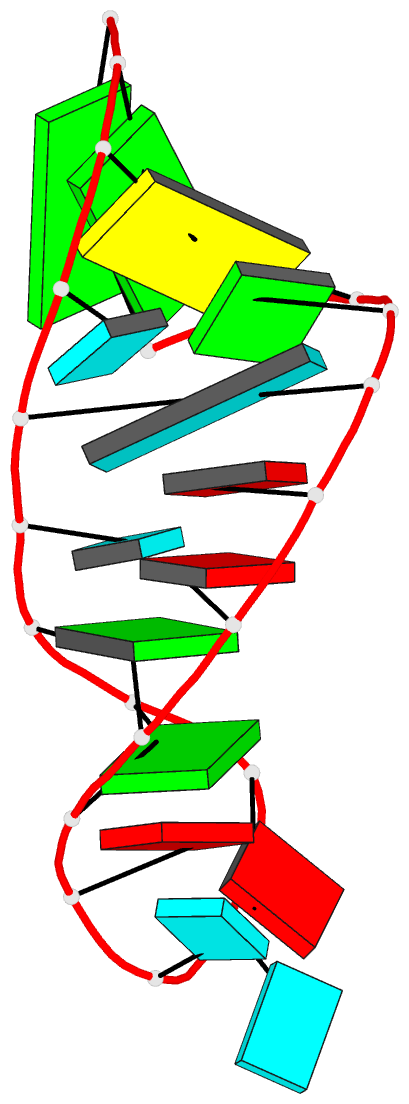
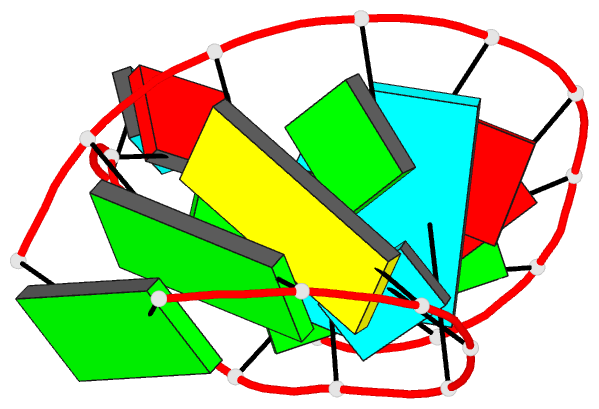
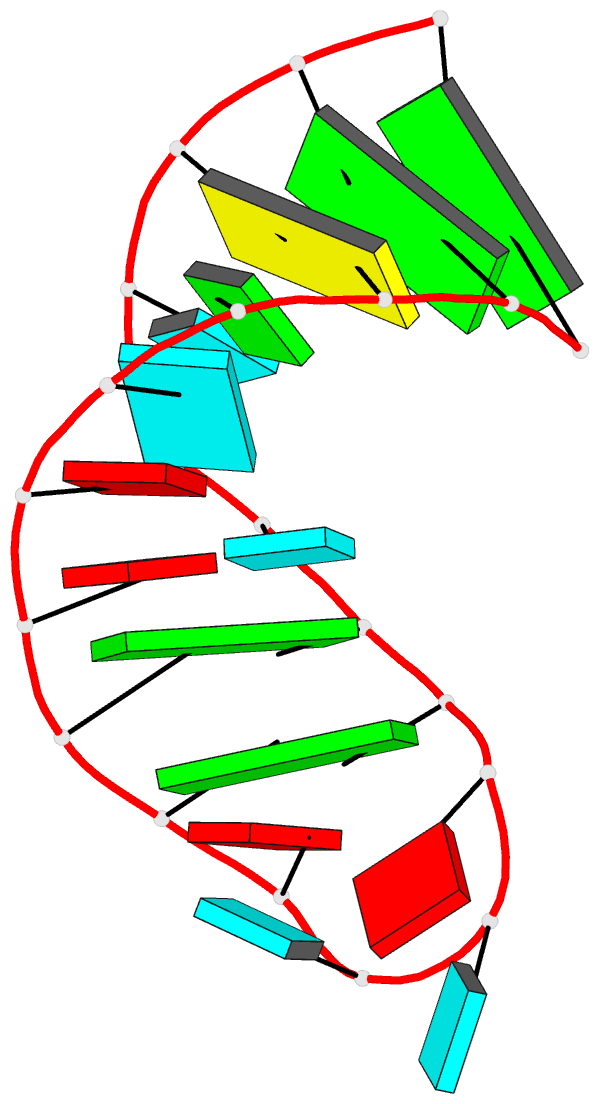
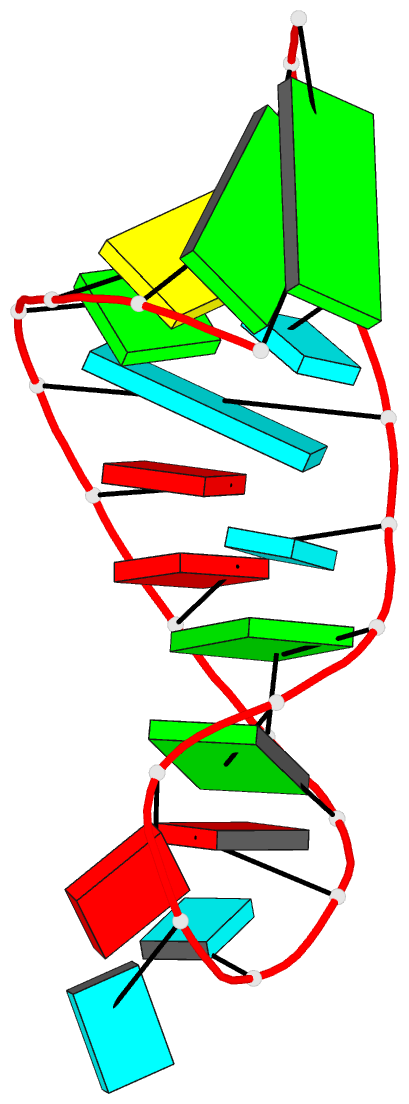
Summary information and primary citation
- PDB-id
-
17ra;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- NMR
- Summary
- Branchpoint helix from yeast and binding site for phage
ga-ms2 coat proteins, NMR, 12 structures
- Reference
-
Smith JS, Nikonowicz EP (1998): "NMR
structure and dynamics of an RNA motif common to the
spliceosome branch-point helix and the RNA-binding site
for phage GA coat protein." Biochemistry,
37, 13486-13498. doi: 10.1021/bi981558a.
- Abstract
- The RNA molecules that make up the spliceosome
branch-point helix and the binding site for phage GA coat
protein share a secondary structure motif in which two
consecutive adenine residues occupy the strand opposite a
single uridine, creating the potential to form one of two
different A.U base pairs while leaving the other adenine
unpaired or bulged. During the splicing of introns out of
pre-mRNA, the 2'-OH of the bulged adenine participates in
the transesterification reaction at the 5'-exon and forms
the branch-point residue of the lariat intermediate. Either
adenine may act as the branch-point residue in mammals, but
the 3'-proximal adenine does so preferentially. When bound
to phage GA coat protein, the bulged adenine loops out of
the helix and occupies a binding pocket on the surface of
the protein, forming a nucleation complex for phage
assembly. The coat protein can bind helices with bulged
adenines at either position, but the 3'-proximal site binds
with greater affinity. We have studied this RNA motif in a
21 nucleotide hairpin containing a GA coat protein-binding
site whose four nucleotide loop has been replaced by a more
stable loop from the related phage Ms2. Using heteronuclear
NMR spectroscopy, we have determined the structure of this
hairpin to an overall precision of 2.0 A. Both adenine
bases stack into the helix, and while all available NOE and
coupling constant data are consistent with both possible
A.U base pairs, the base pair involving the 5'-proximal
adenine appears to be the major conformation. The
3'-proximal bulged adenine protonates at unusually high pH,
and to account for this, we propose a model in which the
protonated adenine is stabilized by a hydrogen bond to the
uridine O2 of the A.U base pair. The 2'-OH of the bulged
adenine adopts a regular A-form helical geometry,
suggesting that in order to participate in the splicing
reaction, the conformation of the branch-point helix in the
active spliceosome may change from the conformation
described here. Thus, while the adenine site preferences of
the spliceosome and of phage GA may be due to protein
factors, the preferred adenine is predisposed in the free
RNA to conformational rearrangement involved in formation
of the active complexes.