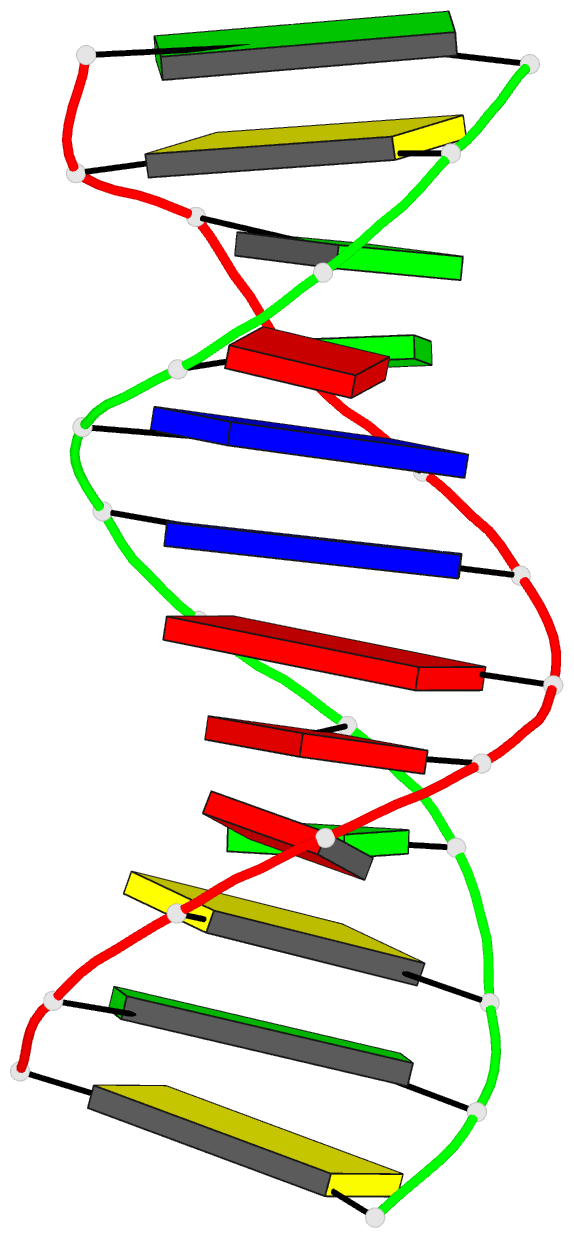
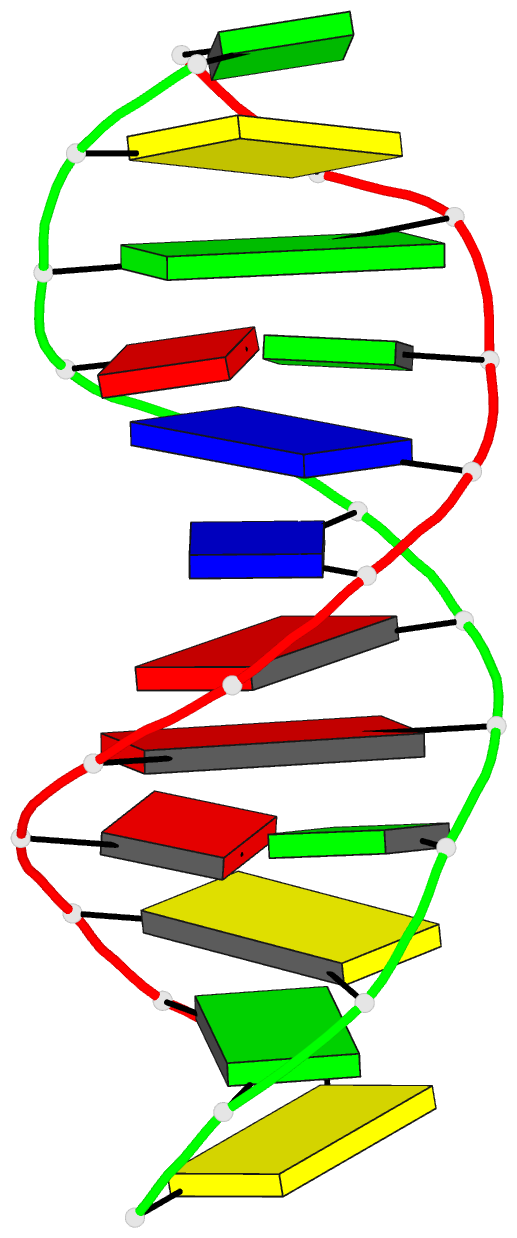
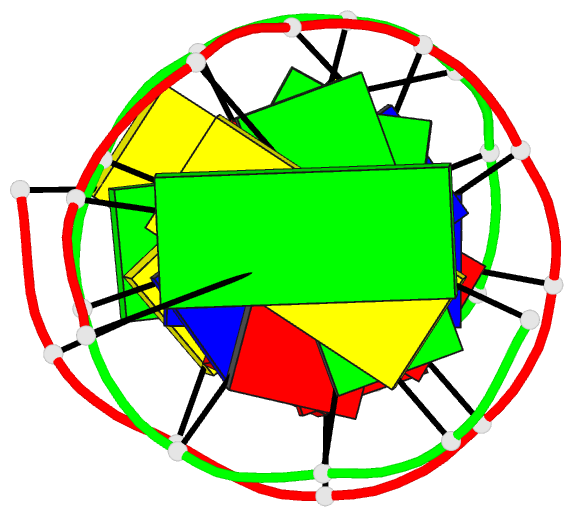
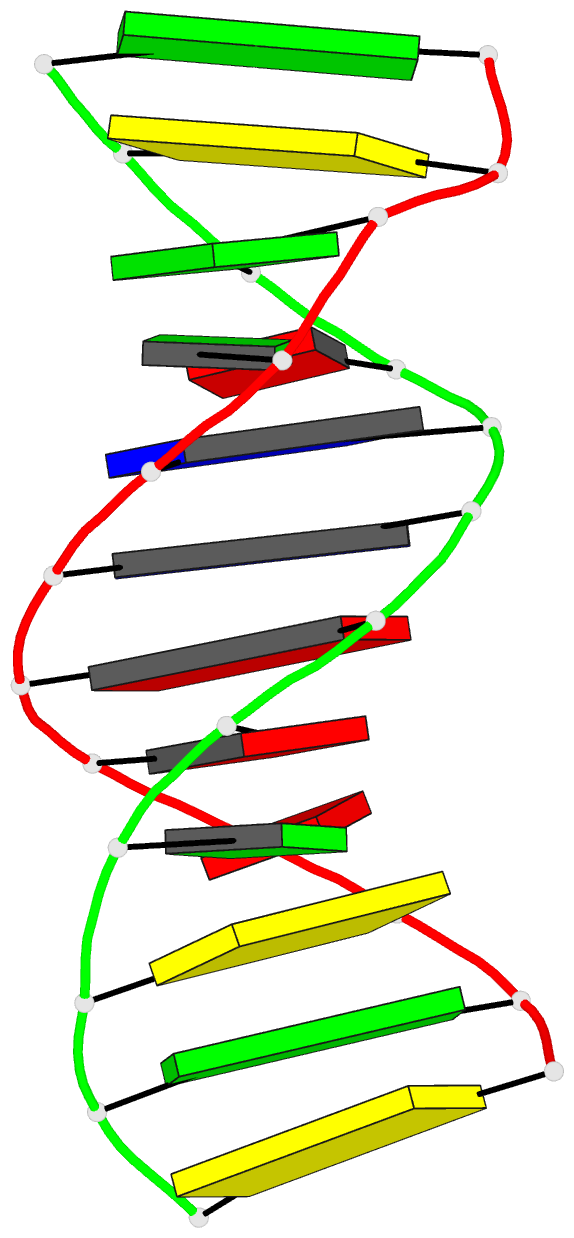
Summary information and primary citation
- PDB-id
-
178d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- X-ray (2.5 Å)
- Summary
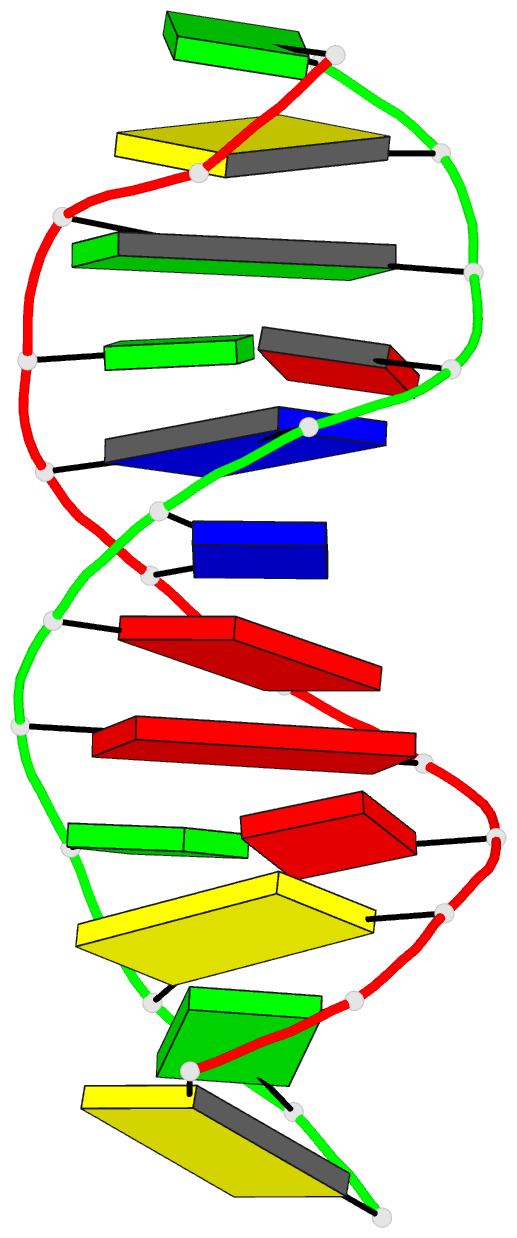
- Crystal structure of a DNA duplex containing
8-hydroxydeoxyguanine.adenine base-pairs
- Reference
-
McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB,
Watson WP, Hunter WN, Brown T (1994): "Crystal
structure of a DNA duplex containing
8-hydroxydeoxyguanine-adenine base pairs."
Biochemistry, 33, 10266-10270.
doi: 10.1021/bi00200a006.
- Abstract
- The crystal structure of the oligonucleotide
d(CGCAAATTO8GGCG), containing the chemically modified base
8-hydroxydeoxyguanine (O8G), has been determined at 2.5-A
resolution and refined to a crystallographic R-factor of
16.8%. The B-type DNA helix contains standard Watson-Crick
base pairs except at the mismatch sites, where O8G adopts a
syn conformation and forms hydrogen bonds to adenine in the
anti conformation. The thermodynamic stability of the
duplex was found by UV melting techniques to be
intermediate between the native oligonucleotide
d(CGCAAATTTGCG) and an oligonucleotide containing A.G
mispairs d(CGCAAATTGGCG). Comparison of the structure of
the O8G(syn).A(anti) base pair with those of Watson-Crick
base pairs has given a reason why O8G.A base pairs are not
well repaired by DNA proofreading enzymes.