Summary information and primary citation
- PDB-id
-
169d;
SNAP-derived features in text and
JSON formats
- Class
- DNA-RNA hybrid
- Method
- NMR
- Summary
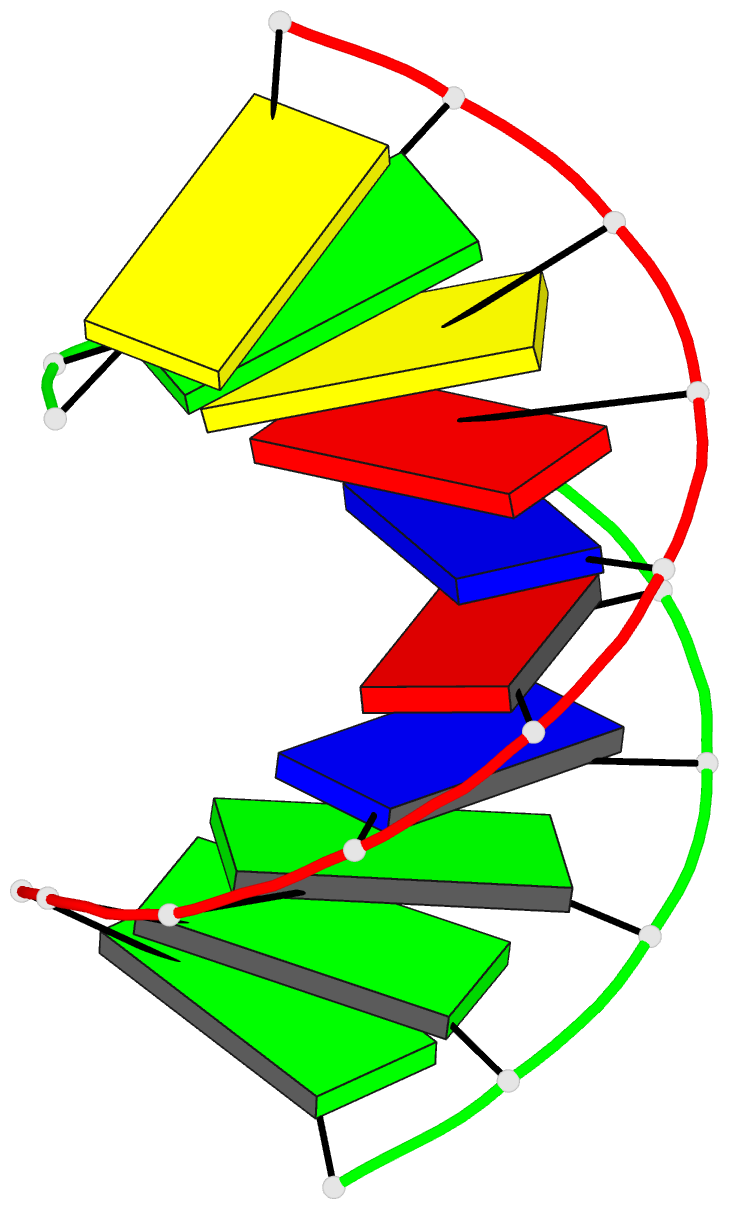
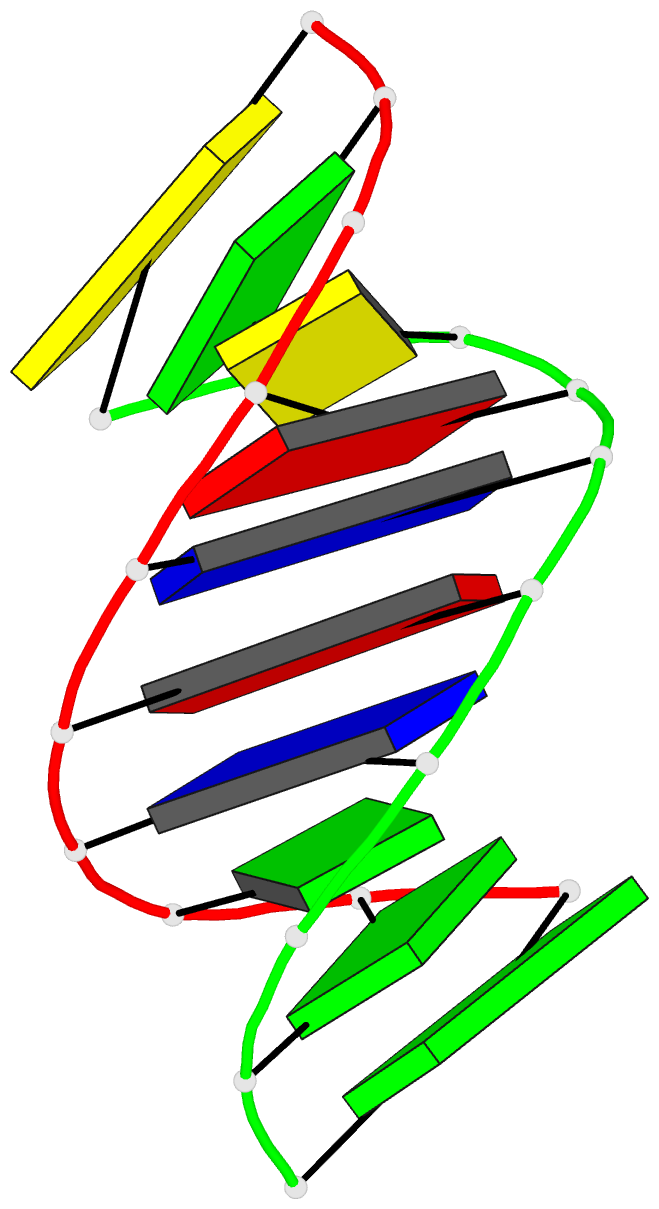
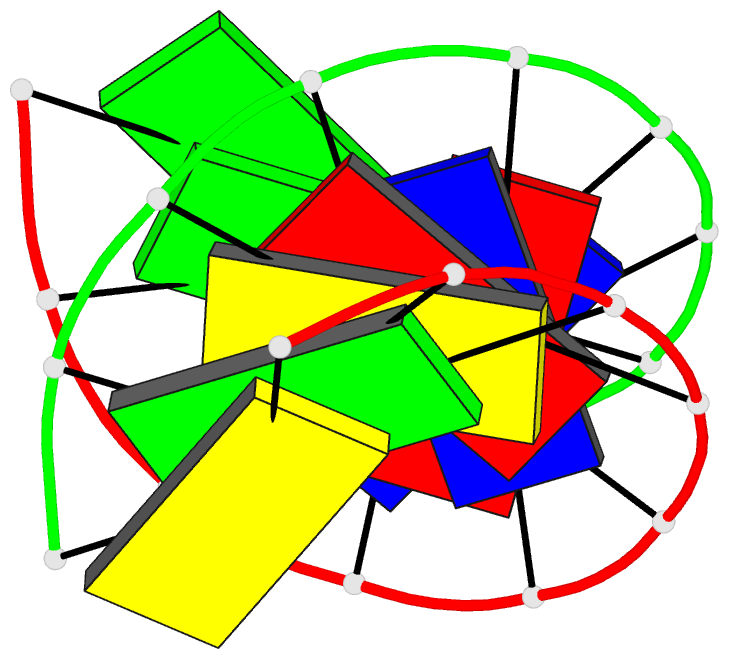
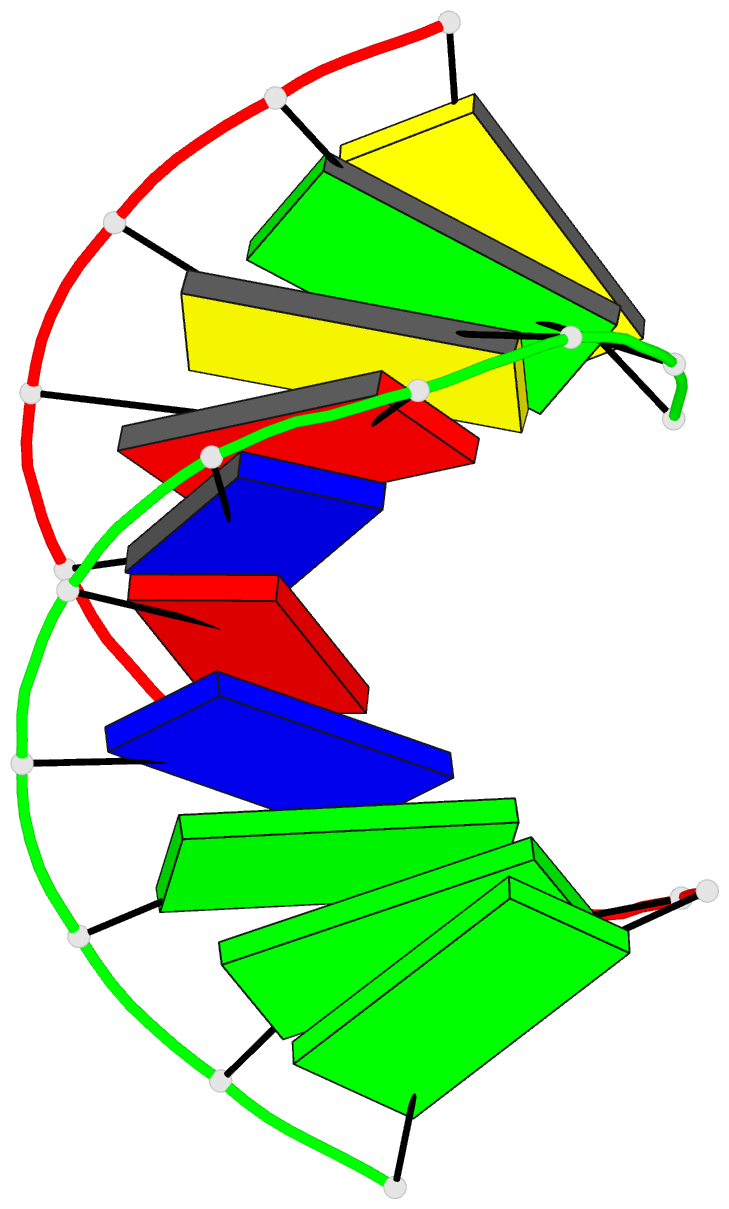
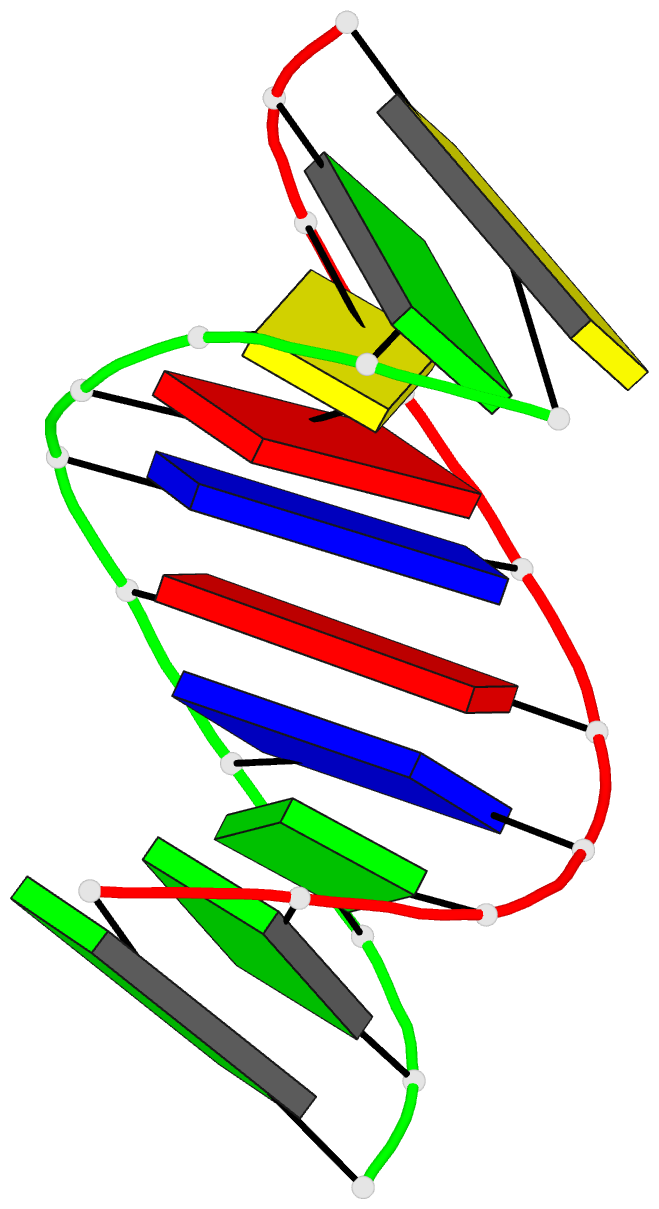
- The solution structure of the
r(gcg)d(tataccc):d(gggtatacgc) okazaki fragment contains
two distinct duplex morphologies connected by a
junction
- Reference
-
Salazar M, Fedoroff OYu, Zhu L, Reid BR (1994): "The
solution structure of the r(gcg)d(TATACCC):d(GGGTATACGC)
Okazaki fragment contains two distinct duplex
morphologies connected by a junction."
J.Mol.Biol., 241, 440-455. doi:
10.1006/jmbi.1994.1519.
- Abstract
- Okazaki fragments are important intermediates in DNA
replication. Chimeric duplexes that are structurally
equivalent to Okazaki fragments also occur during reverse
transcription of RNA retroviruses. Such duplexes consist of
an RNA-DNA chimeric strand base-paired to a pure DNA
strand; hence they have a hybrid duplex "left half"
covalently linked to a "right half" that is pure DNA. We
have determined the solution structure of the synthetic
Okazaki fragment r(gcg)d(TATACCC):d(GGGTATACGC) by means of
two-dimensional NMR, restrained molecular dynamics and full
relaxation matrix simulation of the two-dimensional nuclear
Overhauser effect spectra at various mixing times. The
large negative x-displacement and large positive
inclination in the hybrid section of the duplex are
structural characteristics similar to those found in pure
hybrid duplexes. However, the DNA sugar puckers and the
width and depth of the minor groove in the pure DNA section
are more like B-form DNA, especially beyond the junction.
Thus, this Okazaki fragment duplex assumes a conformation
in solution that is a chimeric mixture of hybrid-form
(H-form) and B-form structures and the overall molecule
cannot be classified as either an A-form or a B-form
duplex. The co-existence of these two different
conformations in a single duplex gives rise to a structural
discontinuity with a bend of approximately 18.1 (+/- 0.4)
degrees at the junction between the hybrid and DNA segments
that may be important for reverse transcriptase binding and
RNase H cleavage of such molecules. Despite the fact that
the solution structure is quite different from the all
A-form structure reported recently for the exact same
molecule in the crystalline state, a surprising number of
local helical parameters were found to be quite similar to
those reported for the crystal structure.