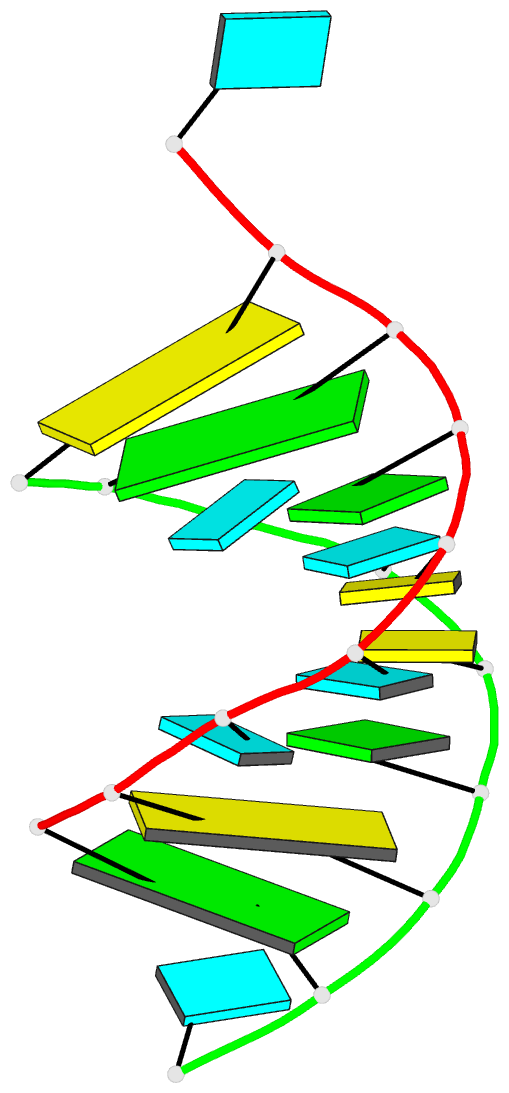
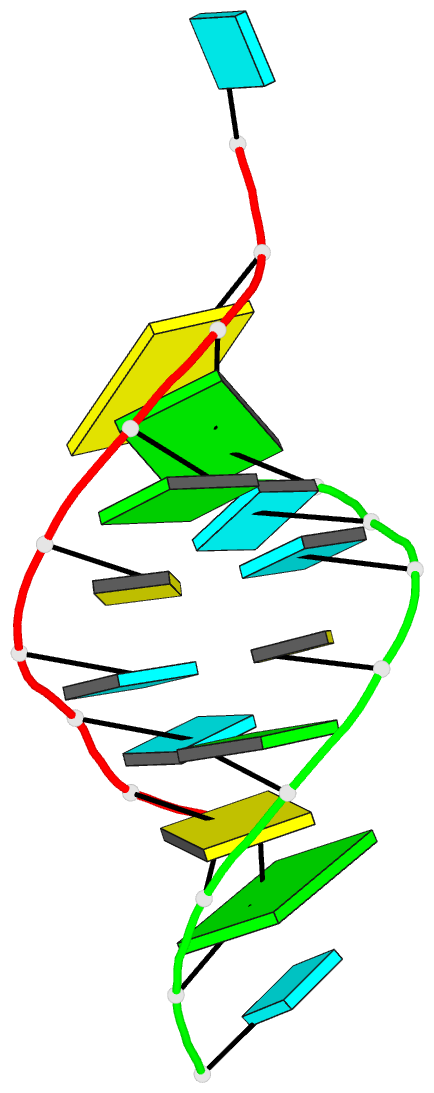
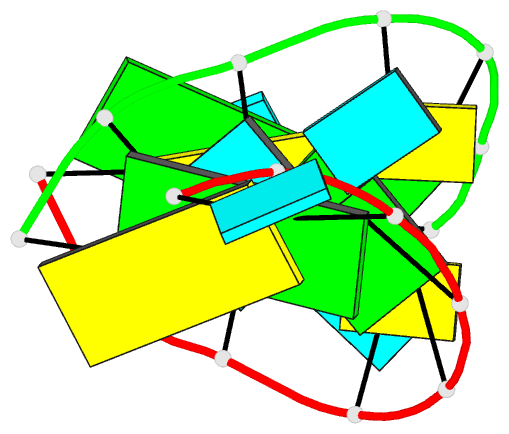
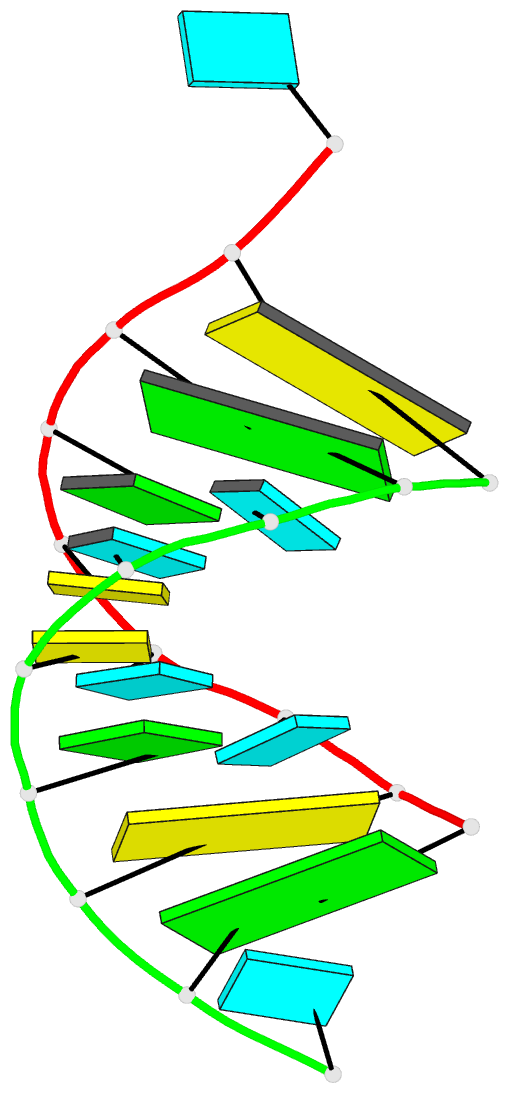
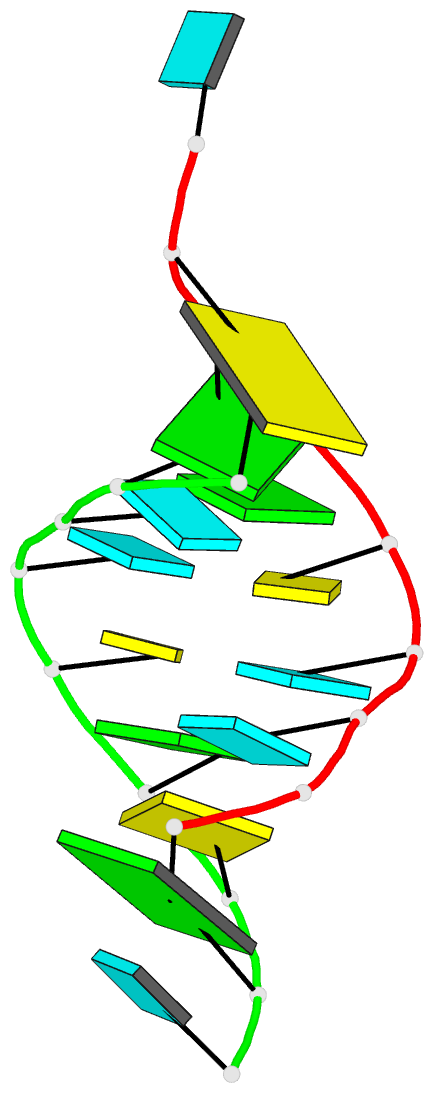
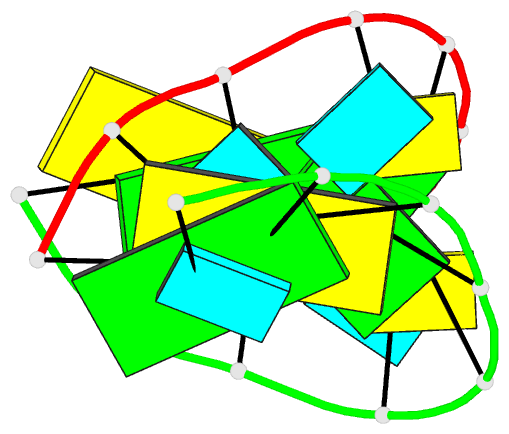
Summary information and primary citation
- PDB-id
-
165d;
SNAP-derived features in text and
JSON formats
- Class
- DNA-RNA hybrid
- Method
- X-ray (1.55 Å)
- Summary
- The structure of a mispaired RNA double helix at 1.6
angstroms resolution and implications for the prediction of
RNA secondary structure
- Reference
-
Cruse WB, Saludjian P, Biala E, Strazewski P, Prange T,
Kennard O (1994): "Structure
of a mispaired RNA double helix at 1.6-A resolution and
implications for the prediction of RNA secondary
structure." Proc.Natl.Acad.Sci.USA,
91, 4160-4164. doi: 10.1073/pnas.91.10.4160.
- Abstract
- The nonamer r(GCUUCGGC)dBrU, where dBrU is
5-bromo-2'-deoxyuridine, contains the tetraloop sequence
UUCG. It crystallizes in the presence of Rh(NH3)6Cl3. In
solution the oligomer is expected to form a hairpin loop
but the x-ray structure analysis, to a resolution of 1.6 A,
indicates an eight-base-pair A-RNA duplex containing a
central block of two G.U and two C.U pairs. Self-pairs
which approximate to Watson-Crick geometry are also formed
in the extended crystal structure between symmetry-related
BrU residues and are part of infinite double-helical
stacks. The G.U pair is a wobble base pair analogous to the
G.T pair found in DNA fragments. The C.U mismatch involves
one hydrogen-bonded contact between the bases and a
bridging water molecule which ensures a good fit of the
base pair in the RNA helix. The BrU.BrU pair is held by two
hydrogen bonds in an orientation which is compatible with
duplex geometry. The structure observed within the crystal
has some parallels with the structure of globular RNAs, and
the presence of stable, noncanonical base pairs has
implications for the prediction of RNA secondary
structure.