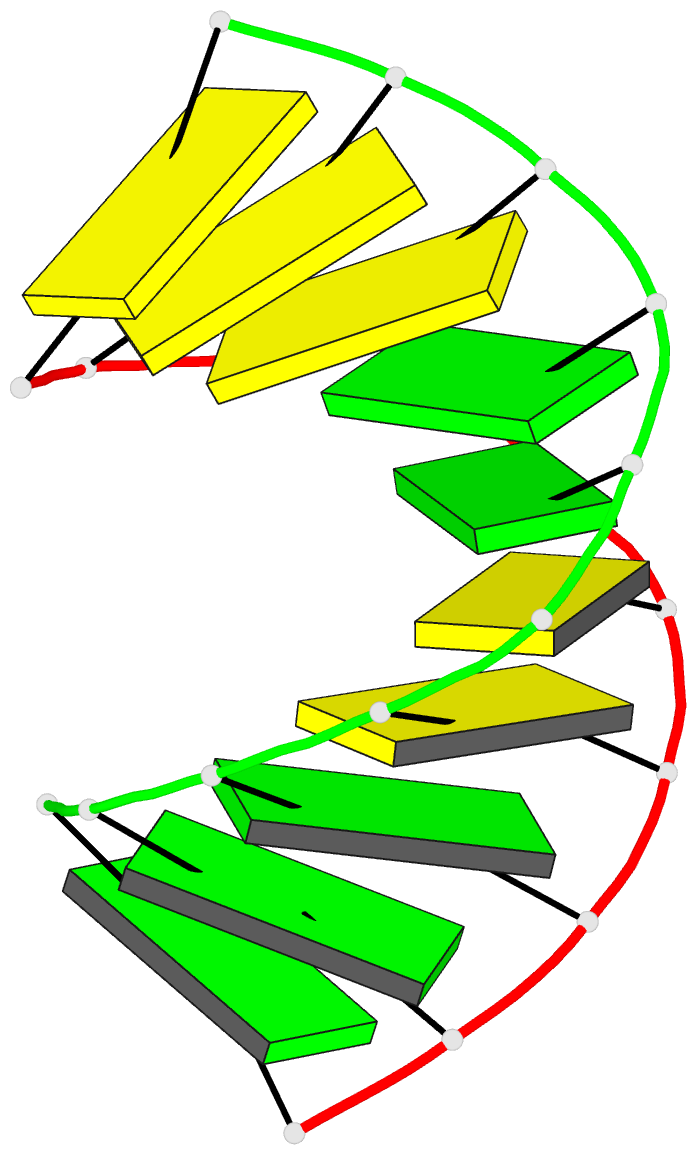
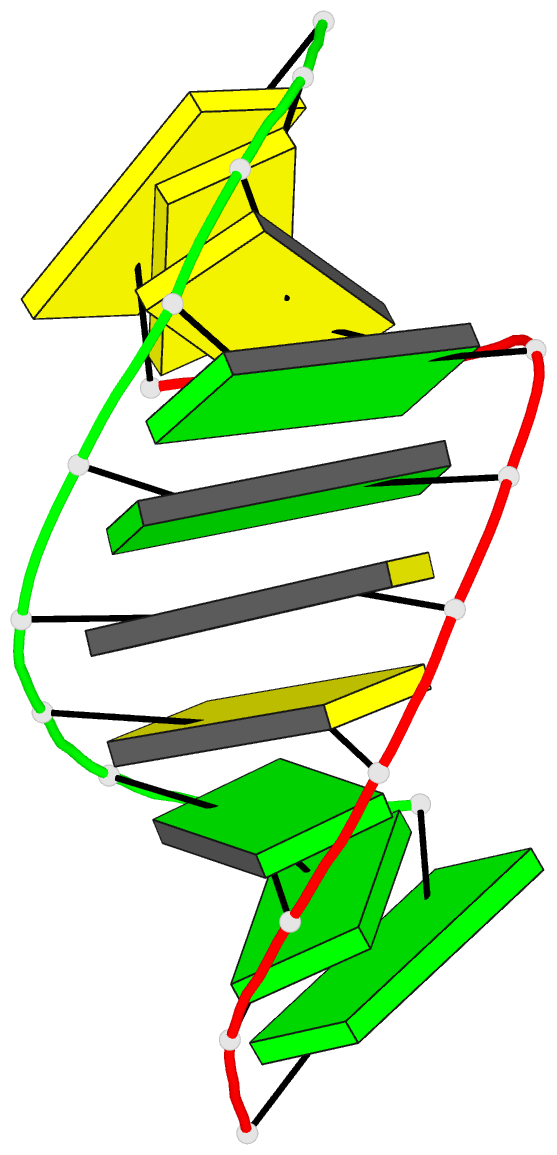
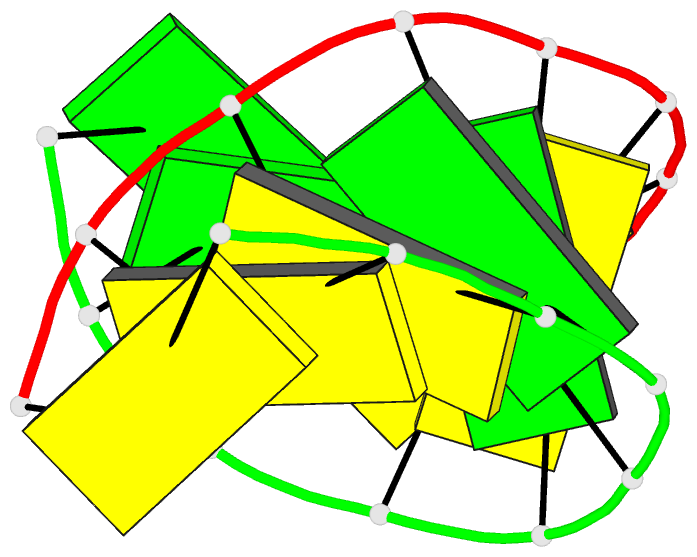
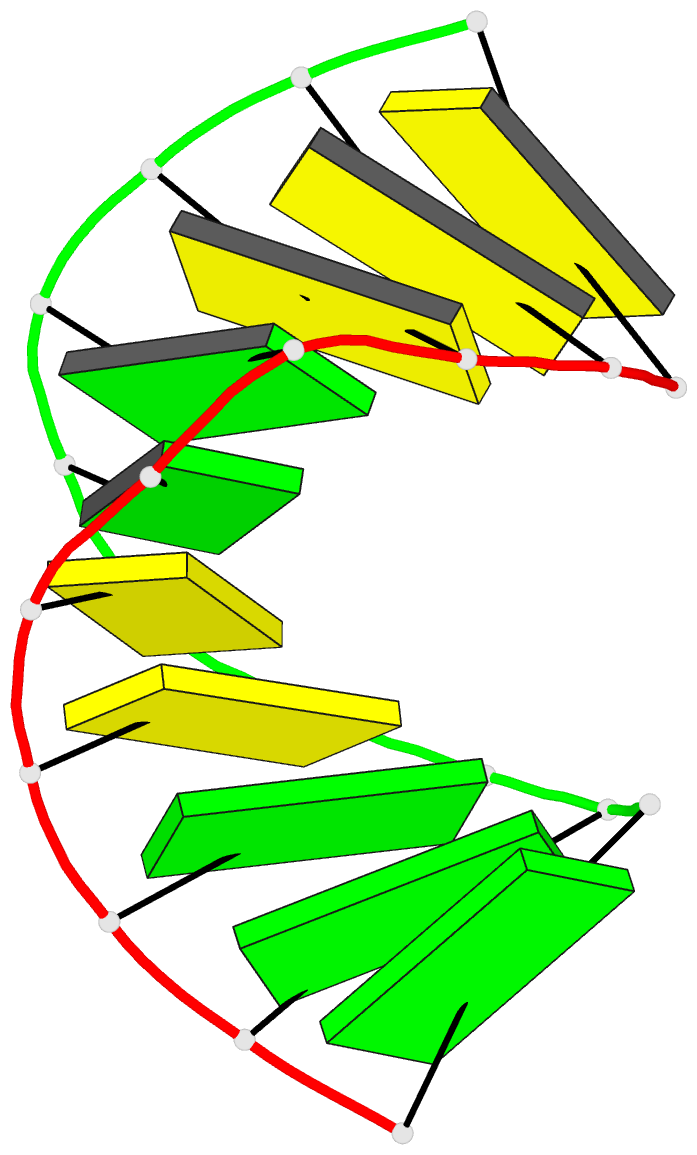
Summary information and primary citation
- PDB-id
-
160d;
SNAP-derived features in text and
JSON formats
- Class
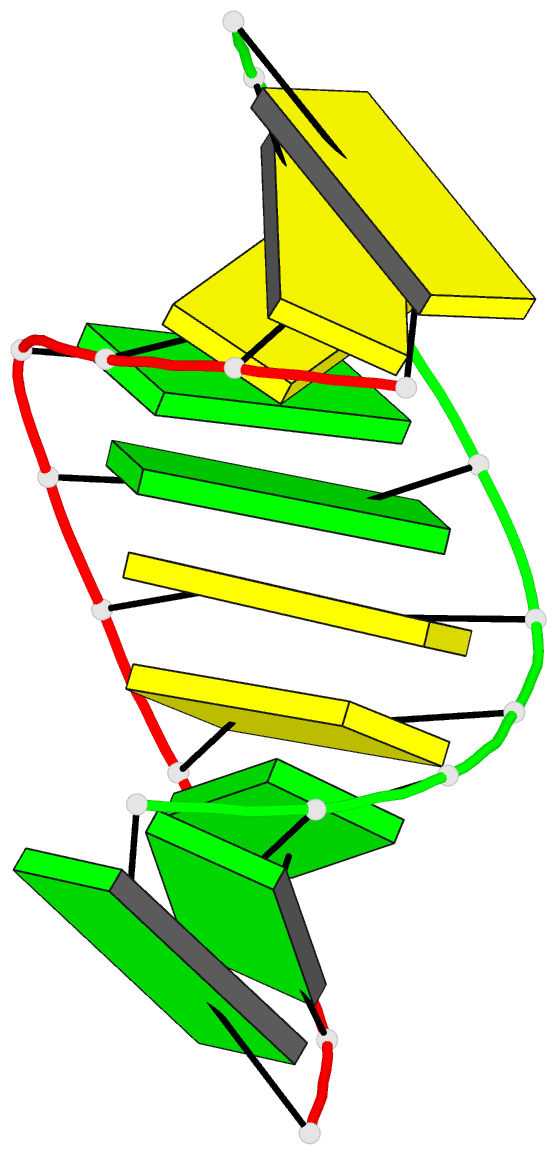
- DNA
- Method
- X-ray (1.65 Å)
- Summary
- High resolution crystal structure of the a-DNA decamer
d(cccggccggg): novel intermolecular base-paired g*(g.c)
triplets
- Reference
-
Ramakrishnan B, Sundaralingam M (1993): "High
resolution crystal structure of the A-DNA decamer
d(CCCGGCCGGG). Novel intermolecular base-paired G*(G.C)
triplets." J.Mol.Biol.,
231, 431-444. doi: 10.1006/jmbi.1993.1292.
- Abstract
- The DNA decamer d(CCCGGCCGGG) crystallizes in the
orthorhombic space group P2(1)2(1)2(1) with a = 24.88, b =
44.60 and c = 46.97 A containing a duplex in the asymmetric
unit. The structure was solved by molecular replacement and
refined to an R factor of 18.5% using 6033 reflections at
1.65 A resolution. The decamer duplex adopts an A-DNA
conformation. The abrupt dislocation of the duplex at the
fourth base-pair G(4).C(17) by an abutting symmetry related
molecule results in distortion of the backbone bonds of the
fifth residue G(5), P-O(5')(alpha) and C(4')-C(5')(gamma),
to the trans conformations from their favored gauche- and
gauche+ conformations, respectively. In this close
encounter the terminal G(10).C(11) base-pair of the
symmetry related molecule hydrogen bonds to the G(4).C(17)
base-pair forming a novel base-paired G(4)*(G10).C(11))
triplet, where G(4) is hydrogen bonded to both G(10) and
C(11). To facilitate this hydrogen bonding the G(4).C(17)
base-pair slides into the minor groove, causing a toll on
the backbone conformation of the adjacent residue G(5). A
similar triplet base-pairing interaction with somewhat
weaker hydrogen bonds occurs at the pseudo dyad related
C(7).G(14) base-pair with G(20) of another symmetry related
duplex. This pseudo triplet interaction (C(7).G(14))*G(20),
does not perturb the backgone alpha and gamma torsions of
G(15). Both the novel base triplets are non-planar. The
abrupt dislocation/bend at the G(4).C(17) base-pair jolts
the global helical base-pair parameters, inclination, tilt,
roll, tip, etc. quite markedly. Therefore a better
description of the helix parameters is obtained by
splitting the duplex and calculating the local helix axis
for the top half consisting of the first three base-pairs,
and the lower half consisting of the last six base-pairs,
omitting the fourth base-pair. The two half duplexes are
bent by only 10 degrees. This structure further
demonstrates that crystal packing interactions, which can
also be governed by base sequence, play a dominant role in
determining DNA conformation.