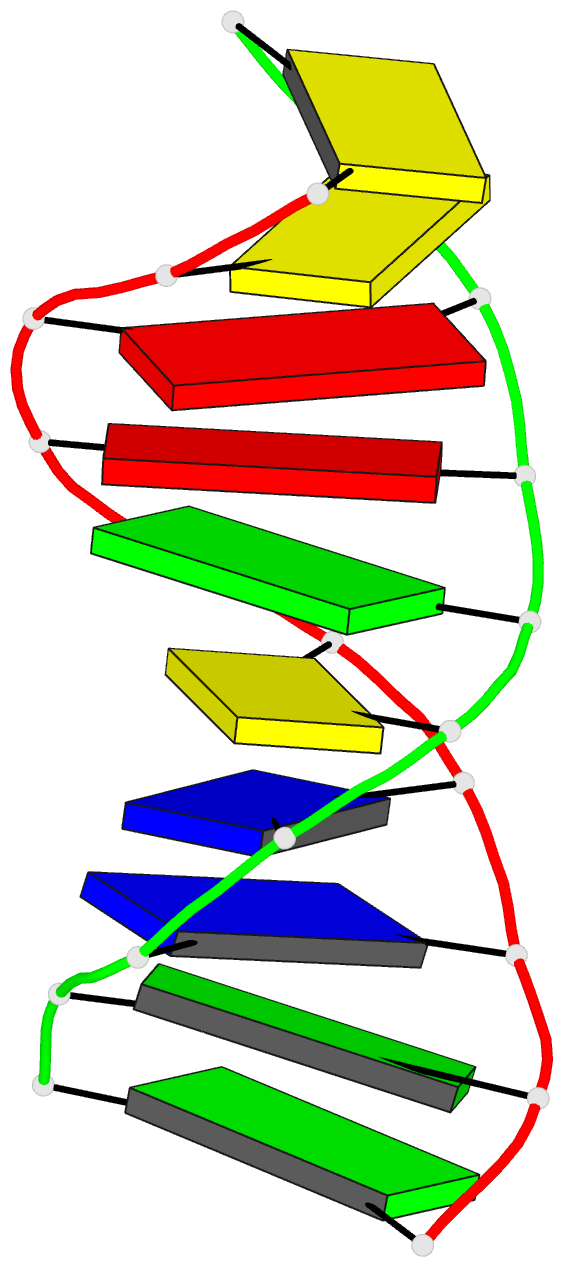
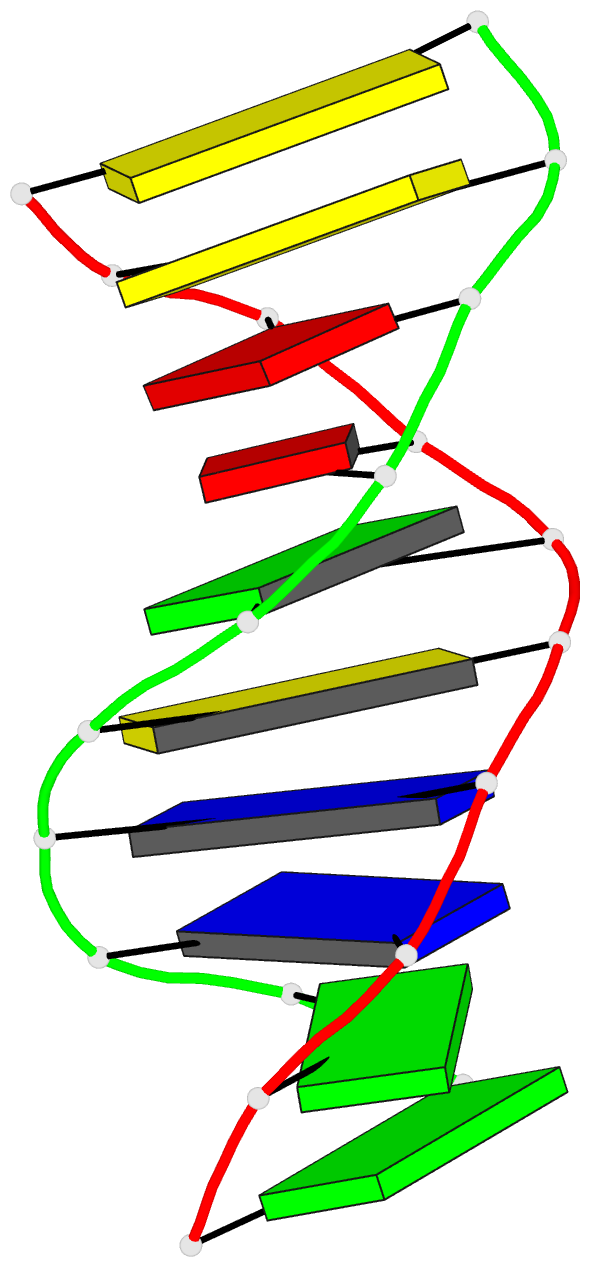
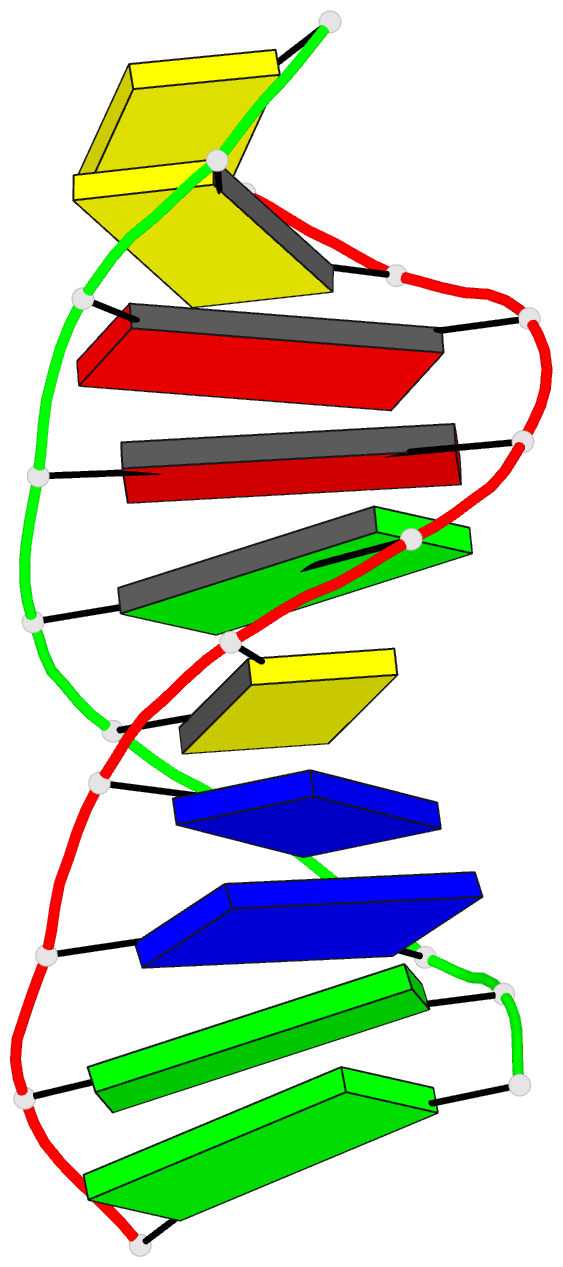
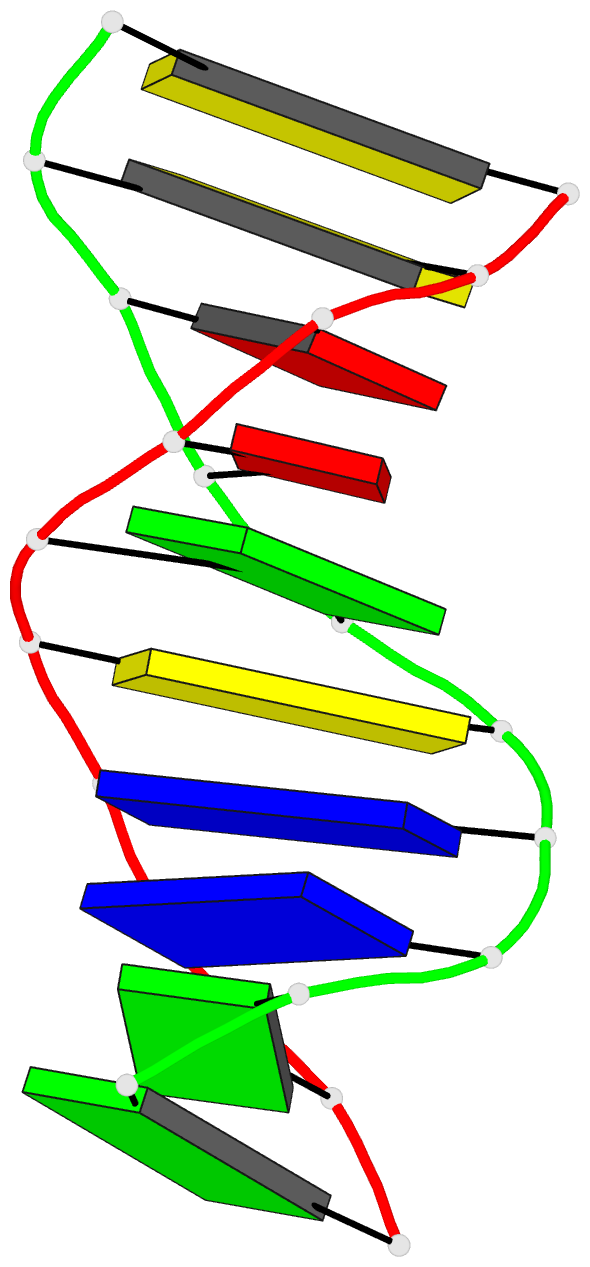
Summary information and primary citation
- PDB-id
-
158d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- X-ray (1.9 Å)
- Summary
- Crystallographic analysis of c-c-a-a-g-c-t-t-g-g and
its implications for bending in b-DNA
- Reference
-
Grzeskowiak K, Goodsell DS, Kaczor-Grzeskowiak M, Cascio
D, Dickerson RE (1993): "Crystallographic
analysis of C-C-A-A-G-C-T-T-G-G and its implications for
bending in B-DNA." Biochemistry,
32, 8923-8931. doi: 10.1021/bi00085a025.
- Abstract
- Stacked B-DNA double helices of sequence
C-C-A-A-G-C-T-T-G-G exhibit the same 23 degrees bend at
-T-G-G C-C-A- across the nonbonded junction between helices
that is observed in the middle of the decamer helix of
sequence C-A-T-G-G-C-C-A-T-G, even though the space group
(hexagonal vs orthorhombic), crystal packing, and
connectedness at the center of the bent segment are quite
different. An identical bend occurs across the interhelix
junction of every monoclinic crystal structure of sequence
C-C-A-x-x-x-x-T-G-G, suggesting that T-G-G-C-C-A
constitutes a natural bending element in B-DNA. The bend
occurs by rolling stacked base pairs about their long axes;
there is no "tilt" component. Of the three possible models
for A-tract bending--bent-A-tract, junction bends, or
bent-non-A--which cannot be distinguished by solution
measurements, all crystallographic evidence over the past
10 years unanimously supports the non-A regions as the
actual bending loci.