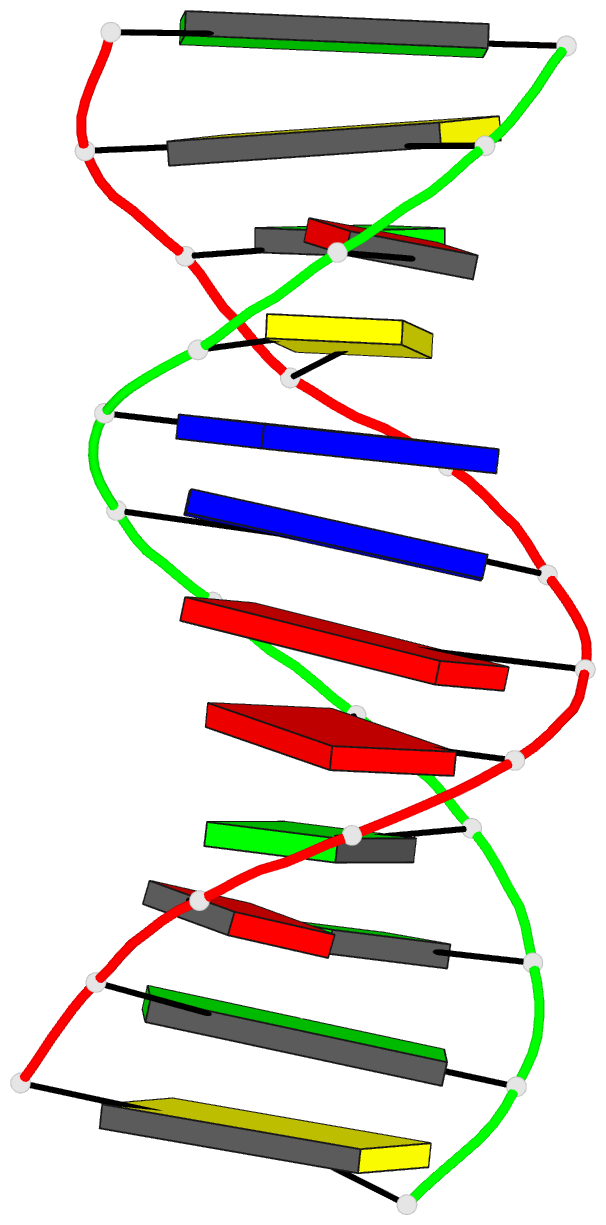
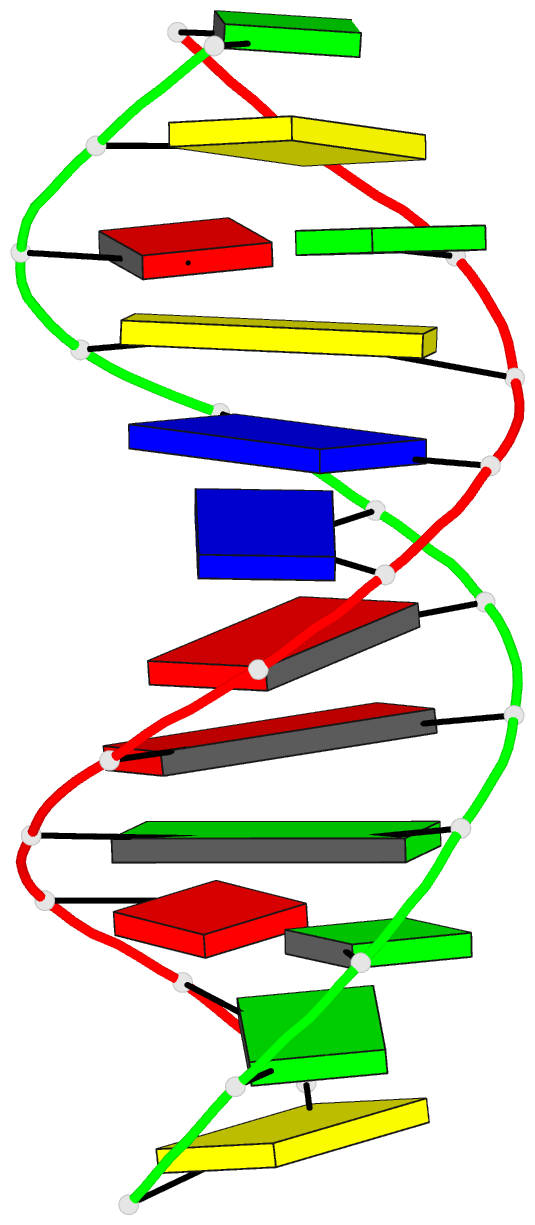
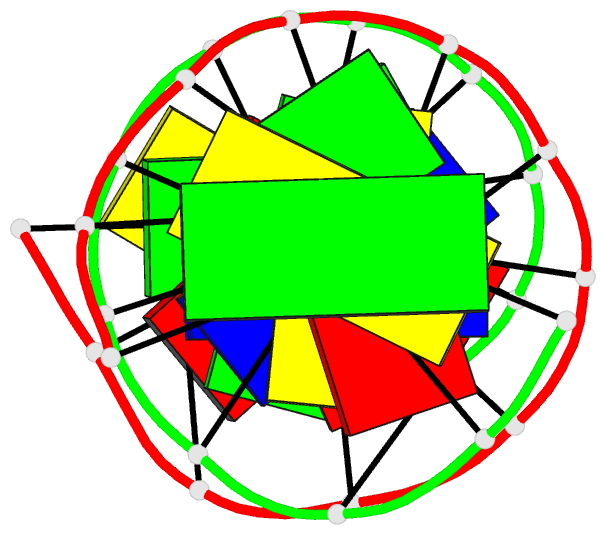
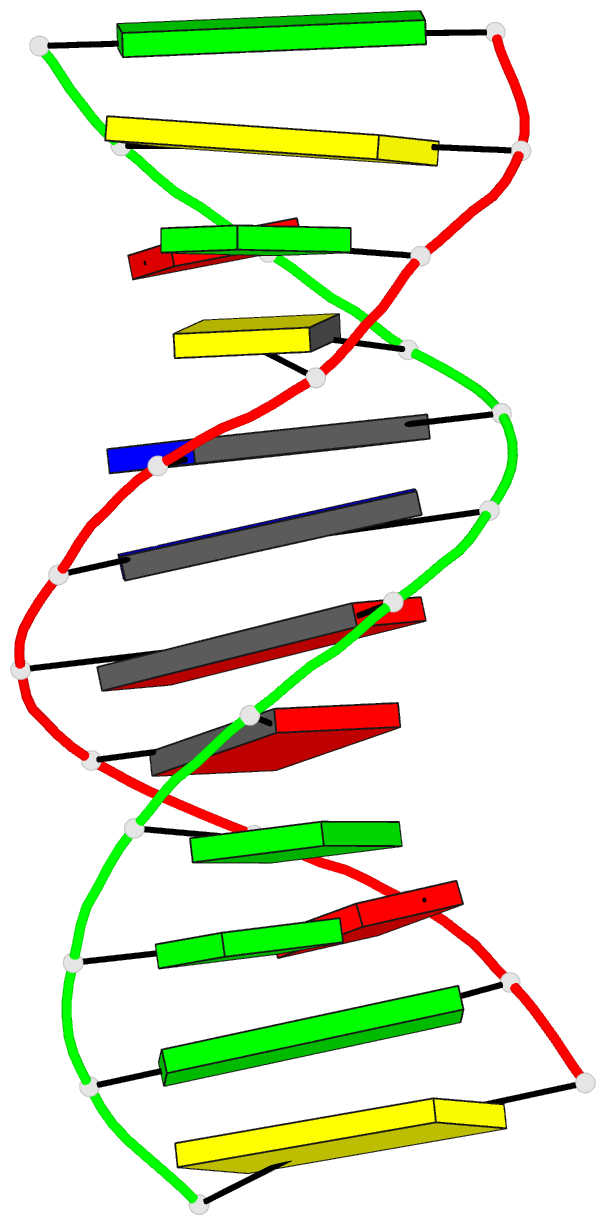
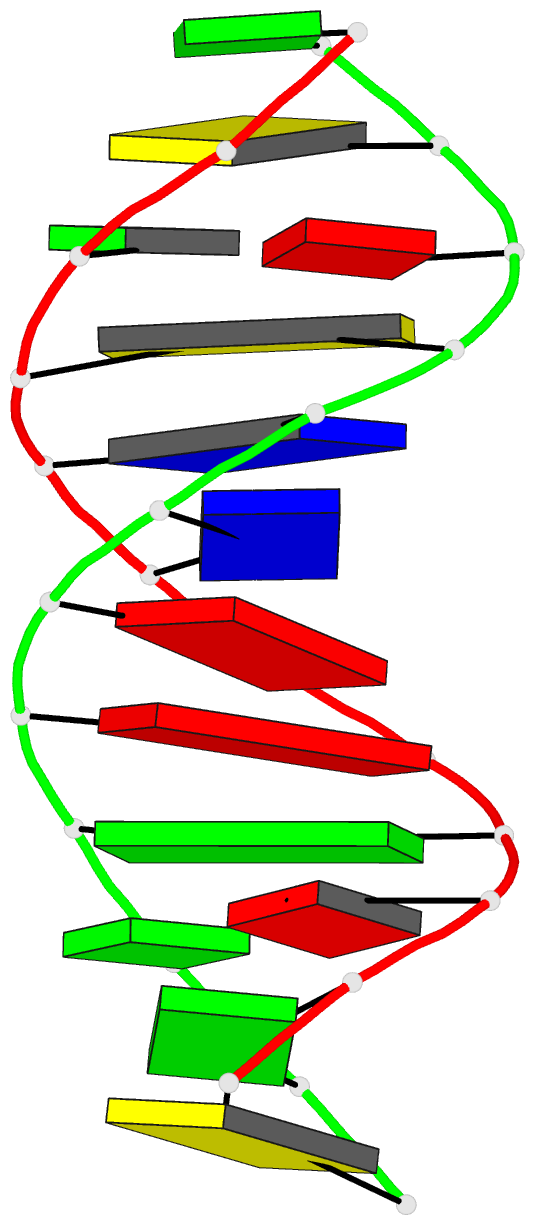
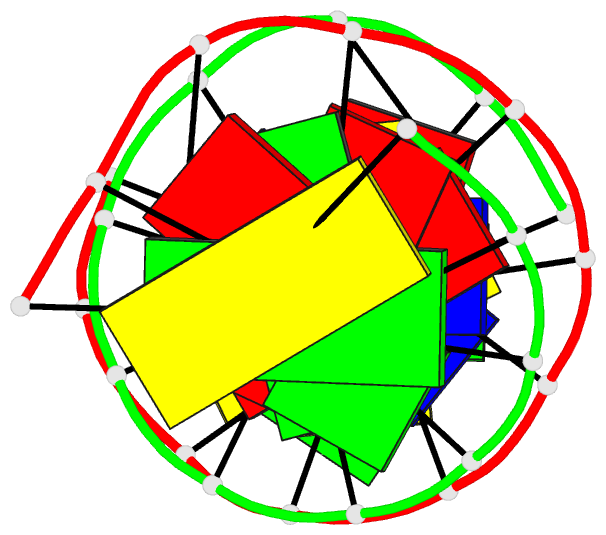
Summary information and primary citation
- PDB-id
-
153d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- X-ray (2.9 Å)
- Summary
- Crystal structure of a mispaired dodecamer,
d(cgagaattc(o6me)gcg)2, containing a carcinogenic
o6-methylguanine
- Reference
-
Ginell SL, Vojtechovsky J, Gaffney B, Jones R, Berman HM
(1994): "Crystal
structure of a mispaired dodecamer,
d(CGAGAATTC(O6Me)GCG)2, containing a carcinogenic
O6-methylguanine." Biochemistry,
33, 3487-3493. doi: 10.1021/bi00178a004.
- Abstract
- The crystal structure of the synthetic deoxydodecamer
d(CGAGAATTC(O6Me)GCG)2 has been determined and refined to
an R-factor of 16.9% with data up to 2.9-A resolution. This
sequence contains two mismatched base pairs between
O6-methylguanine and adenine with the arrangement
A(syn).(O6-Me)G(anti) which differs from the geometry
observed in solution by NMR. The intermolecular arrangement
is equivalent to the other isomorphous deoxydodecamers.
However, the weakening of some significant crystal packing
contacts was observed and related to the effect of stacking
between the mispaired adenine and the adjacent guanine in
the sequence. The structure is highly hydrated, with a
total of 49 solvent molecules located. The methyl group and
the mismatched base-pair geometry locally disrupt the
B-DNA-type solvent network with two solvent molecules found
close to the N1 and N6 of the mispaired adenine.