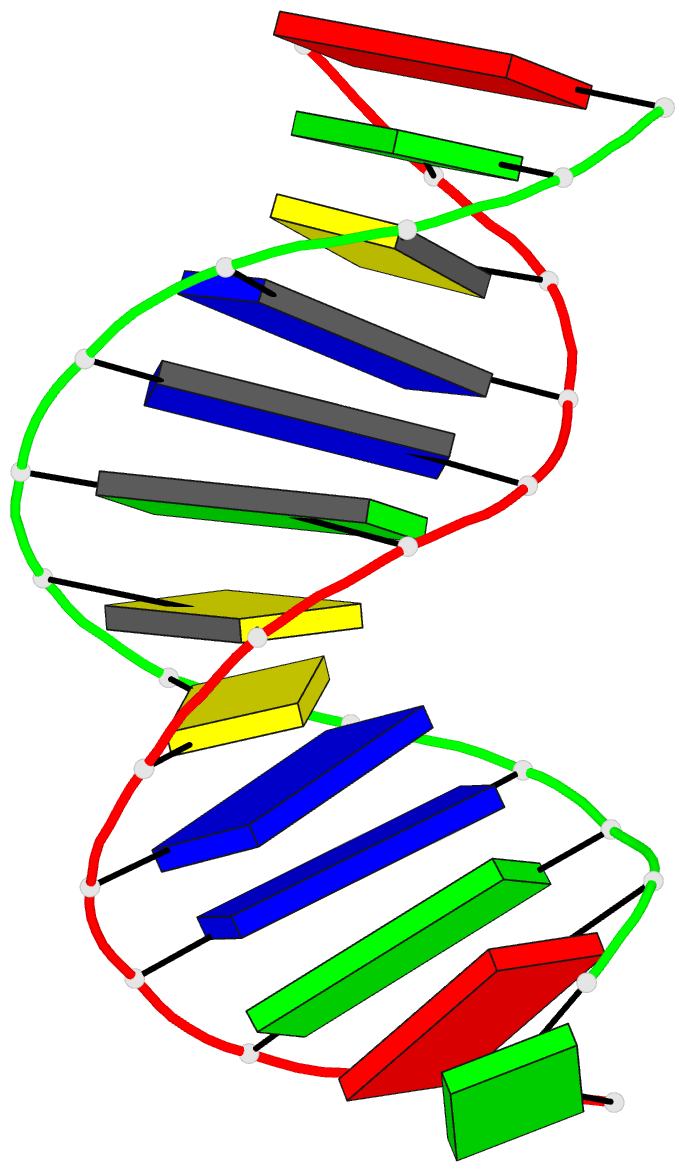
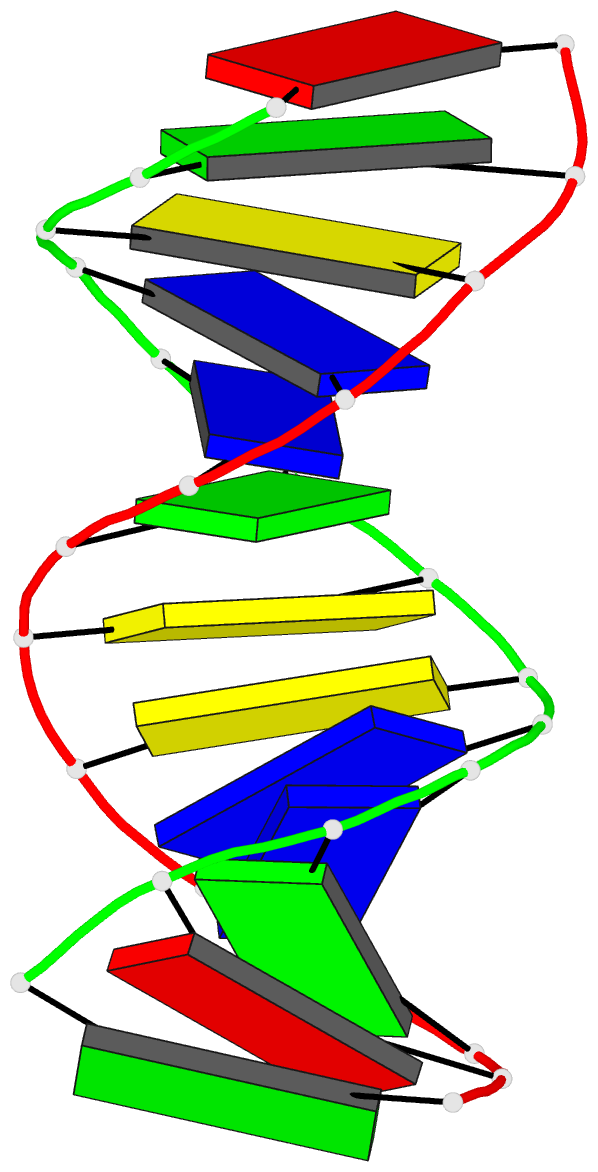
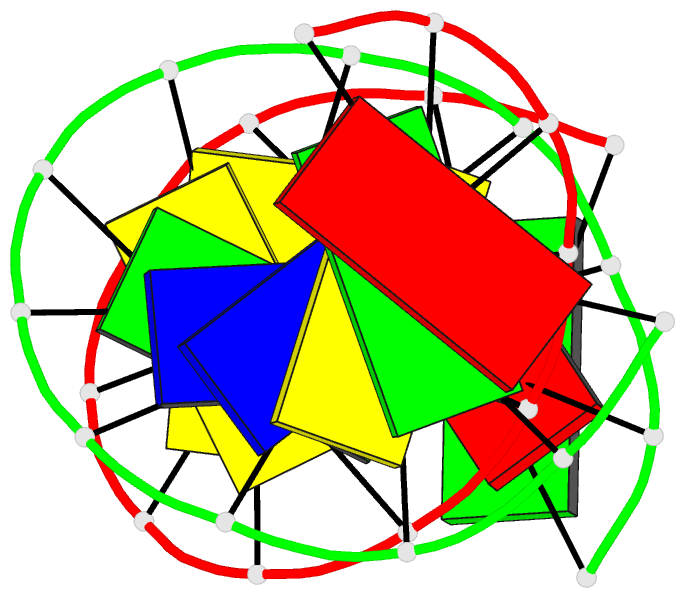
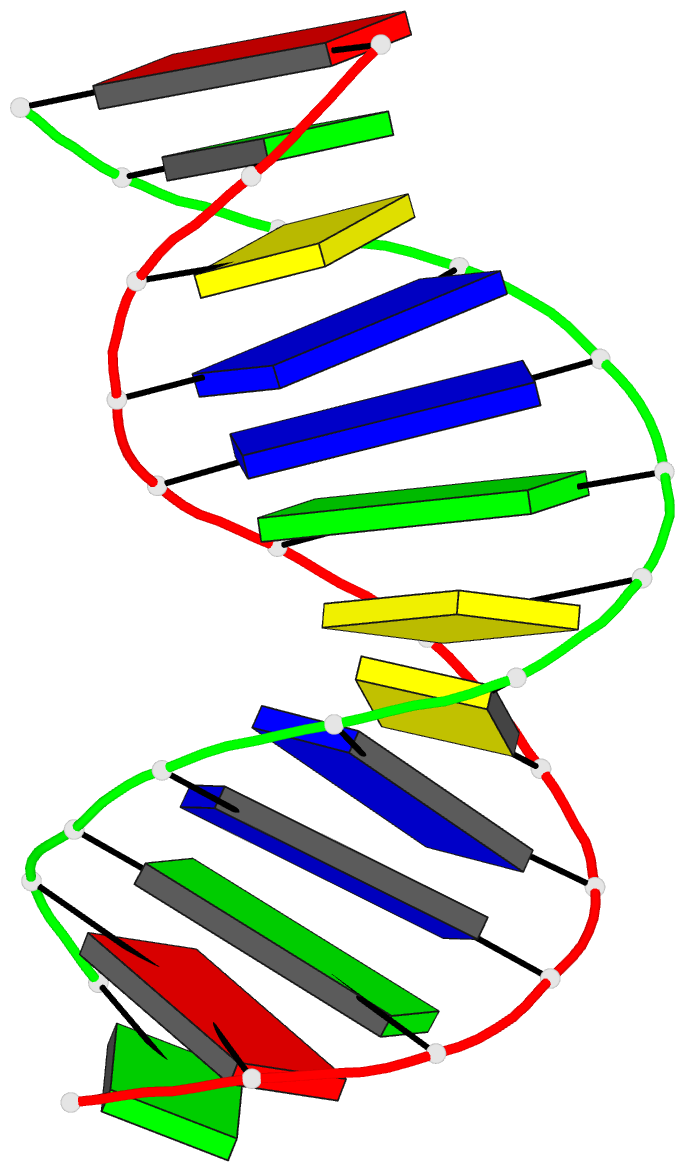
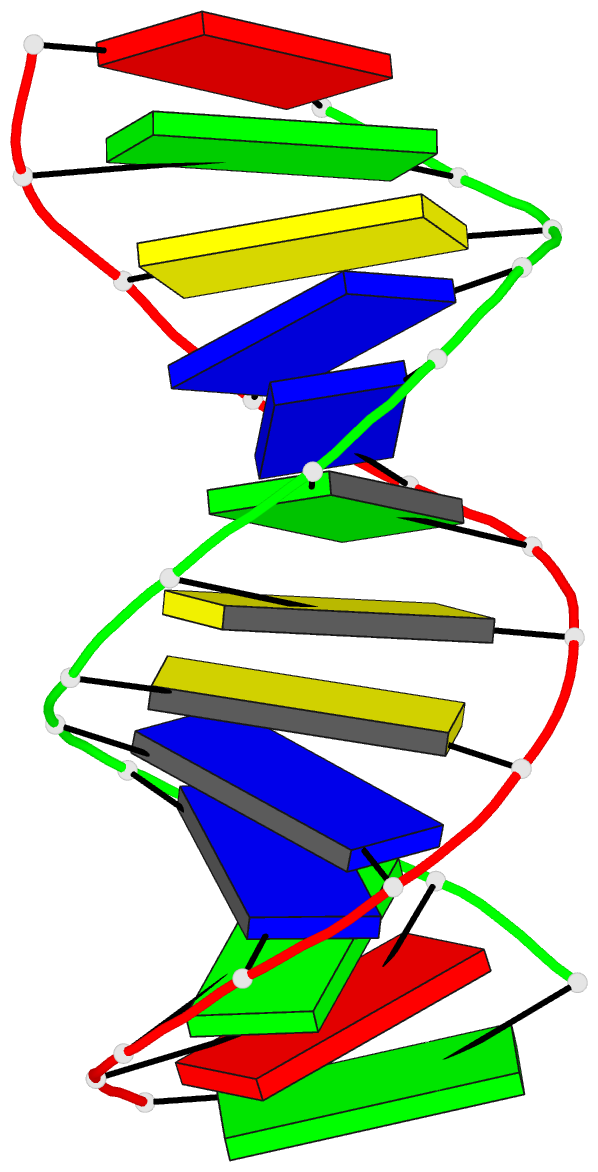
Summary information and primary citation
- PDB-id
-
142d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of a conserved DNA sequence from the
hiv-1 genome: restrained molecular dynamics simulation with
distance and torsion angle restraints derived from
two-dimensional NMR spectra
- Reference
-
Mujeeb A, Kerwin SM, Kenyon GL, James TL (1993):
"Solution
structure of a conserved DNA sequence from the HIV-1
genome: restrained molecular dynamics simulation with
distance and torsion angle restraints derived from
two-dimensional NMR spectra." Biochemistry,
32, 13419-13431. doi: 10.1021/bi00212a007.
- Abstract
- The three-dimensional solution structure of a
trisdecamer DNA duplex sequence,
d(AGCTTGCCTTGAG).d(CTCAAGGCAAGCT), from a conserved region
of HIV-1 genome's long terminal repeat, has been
investigated using NMR spectroscopy and restrained
molecular dynamics calculations. Interproton distances
derived from two-dimensional nuclear Overhauser enhancement
(2D NOE) experiments, using the iterative complete
relaxation matrix algorithm MARDIGRAS, and torsion angles
for sugar rings, estimated from stimulated fitting of
double-quantum-filtered correlation (2QF-COSY) spectra,
were obtained [Mujeeb, A., Kerwin, S. M., Egan, W. M.,
Kenyon, G. L., & James, T. L. (1992) Biochemistry 31,
9325-38]. These structural restraints have now been
employed as the basis for structure refinement using
restrained molecular dynamics (rMD) to search
conformational space for structures consistent with the
experimental restraints. Specifically, upper and lower
bounds on the restraints were incorporated into the AMBER
(version 4.0) total potential energy function of the
system, the bounds being used to define the width of a
flat-well penalty term in the AMBER force field. Confidence
in the time-averaged structure obtained is engendered by
convergence to essentially the same structure
(root-mean-square deviation approximately 0.9 A) when two
quite different DNA models, A-DNA and B-DNA (RMSD
approximately 6.5 A), were employed as starting structures
and when various initial trajectories were used for the rMD
runs. The derived structure is further supported by the
total energy calculated, the restraint violation energy,
the restraint deviations, and the fit with experimental
data. For the latter, the sixth-root residual index
indicated a good fit of the determined structure with
experimental 2D NOE spectral intensities (R1x < 0.07),
and the RMS difference between vicinal proton coupling
constants calculated for the derived structure and
experimental coupling constants were also in reasonable
agreement (JRMS = 0.9 Hz). While the structure of the
trisdecamer is basically in the B-DNA family, some
structural parameters manifest interesting local
variations. The helix parameters indicate that, compared
with classical B-DNA, the structure is longitudinally more
compressed. Local structural variations at the two TG steps
in particular together create bending into the major groove
of the duplex. Comparison of the two-CTTG-tetrads in the
duplex reveals that they have similar structures, with the
TT moieties being almost identical; however, the -CTTG-pur
sequence has a larger roll and slide for the -TG- step than
for the -CTTG-pyr sequence, in accord with published X-ray
crystallographic conclusions.