Summary information and primary citation
- PDB-id
-
139d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- NMR
- Summary
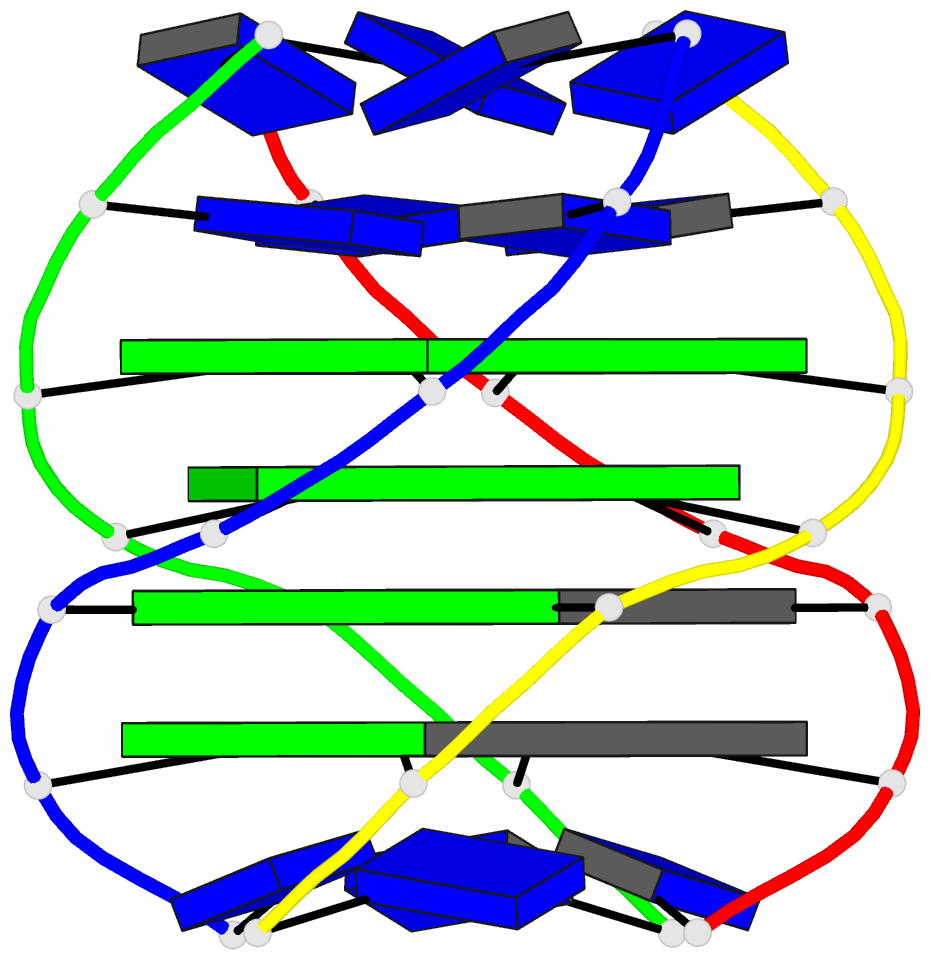
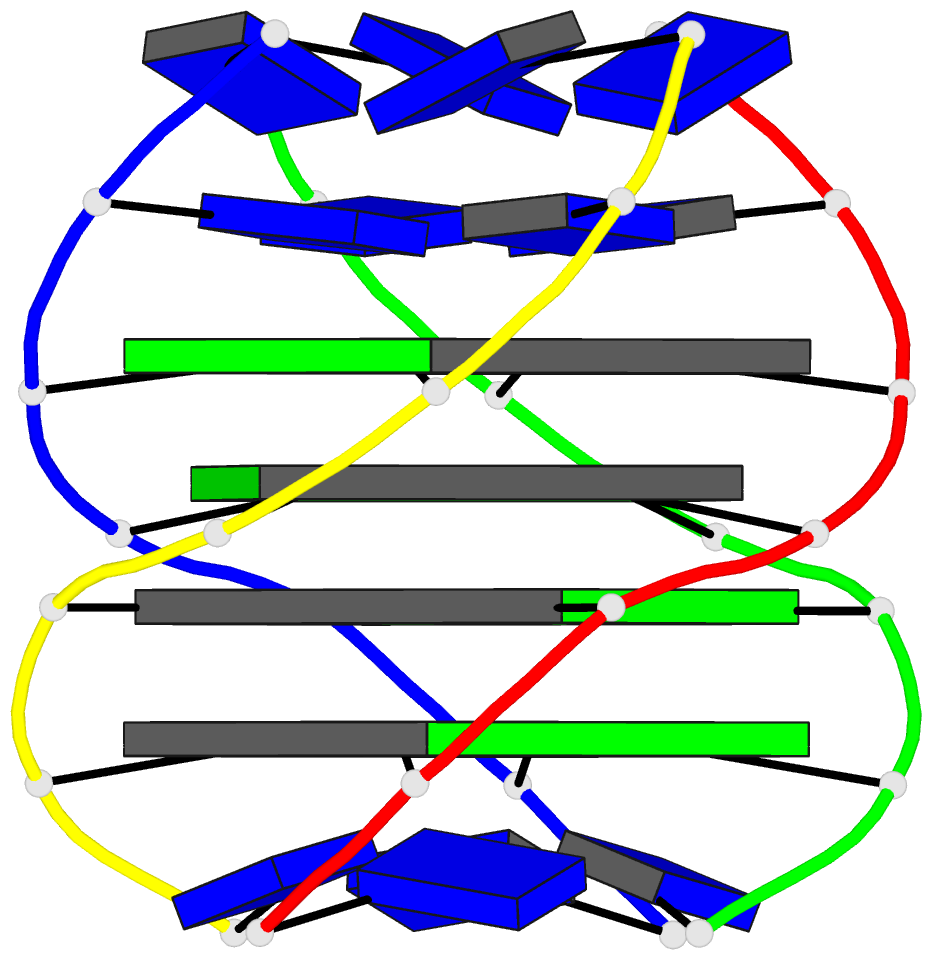
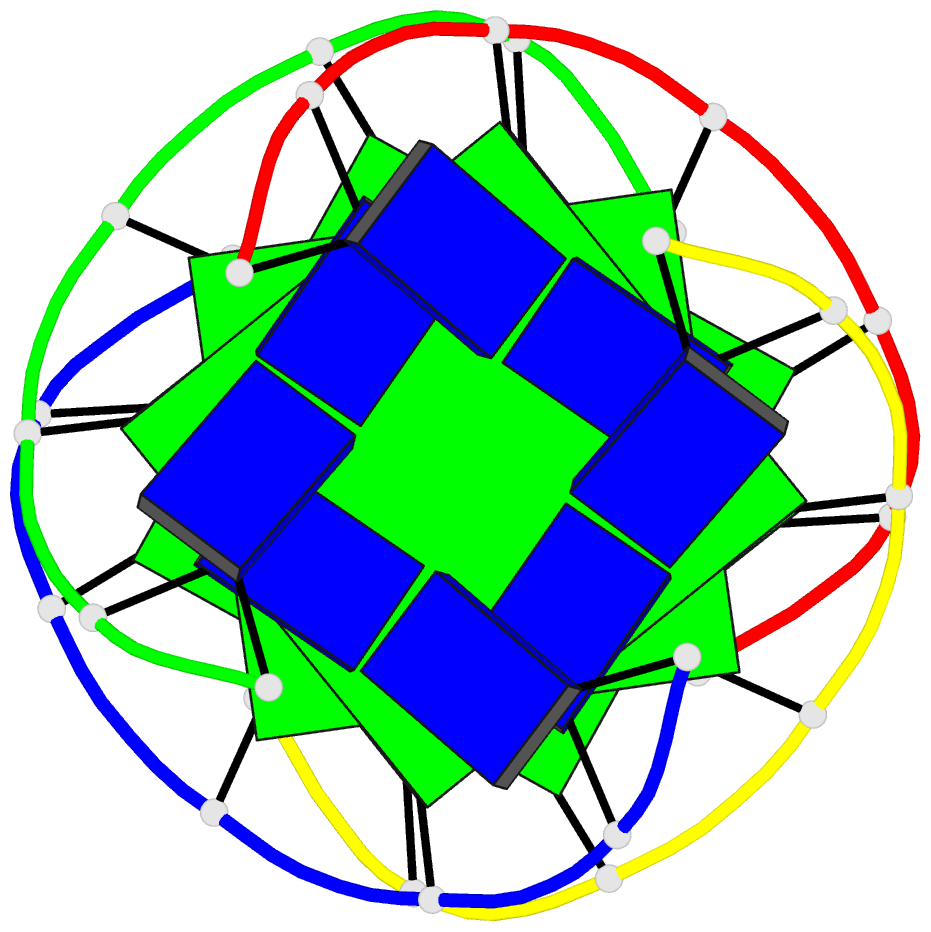
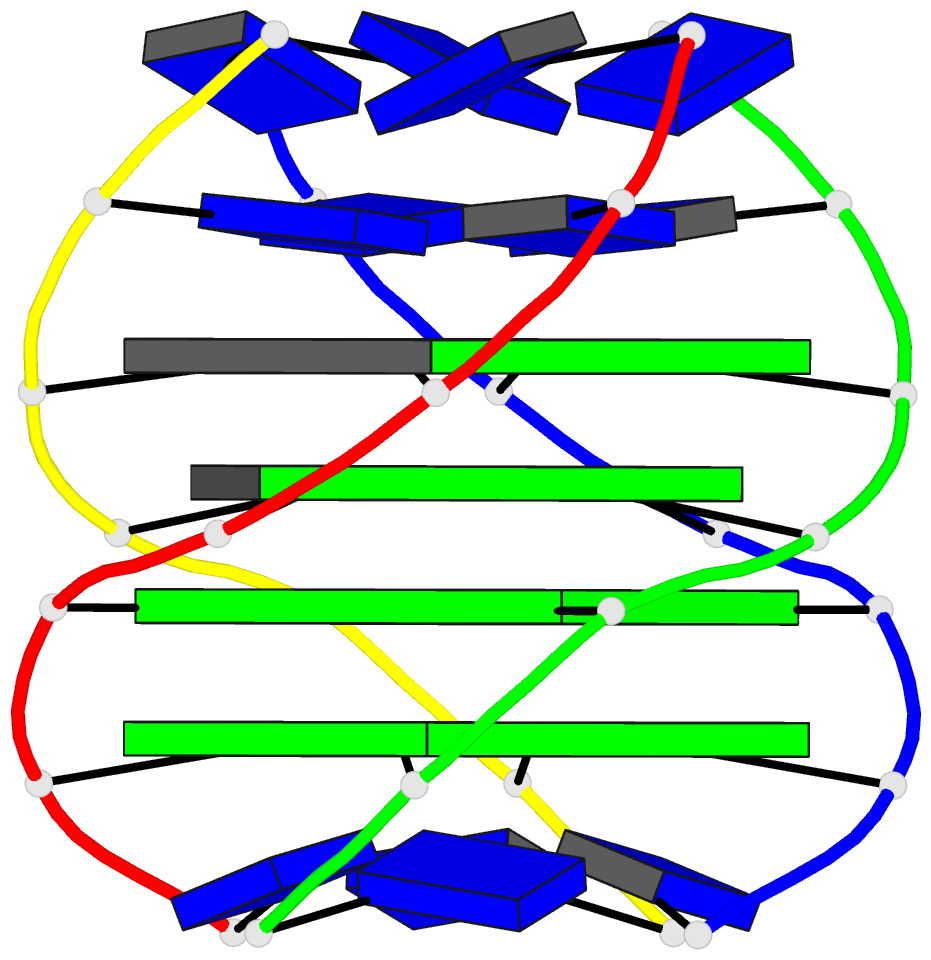
- Solution structure of a parallel-stranded g-quadruplex
DNA
- Reference
-
Wang Y, Patel DJ (1993): "Solution
structure of a parallel-stranded G-quadruplex DNA."
J.Mol.Biol., 234, 1171-1183.
doi: 10.1006/jmbi.1993.1668.
- Abstract
- This paper reports on the solution structure of a
parallel-stranded G-quadruplex formed by the Tetrahymena
telomeric sequence d(T-T-G-G-G-G-T) whose NMR parameters in
potassium cation containing solution were previously
published from our laboratory. The structure was determined
by combining a quantitative analysis of the NMR data with
molecular dynamics calculations including relaxation matrix
refinement. The combined NMR-computational approach yielded
a set of seven distance-refined structures with pairwise
RMSDs ranging from 0.66 to 1.30 A for the central G-G-G-G
tetranucleotide segment. Four of the seven structures were
refined further using complete relaxation-matrix
calculations to yield solution structures with pairwise
RMSDs ranging from 0.64 to 1.04 A for the same
tetranucleotide segment. The R-factors also decreased on
proceeding from the distance-refined to relaxation
matrix-refined structures. The four strands of the
G-quadruplex are aligned in parallel and are related by a
4-fold symmetry axis coincident with the helix axis.
Individual guanines from each strand form planar G.G.G.G
tetrad arrangements with each tetrad stabilized by eight
hydrogen bonds involving the Watson-Crick and Hoogsteen
edges of the guanine bases. All guanines adopt anti
glycosidic torsion angles and S type sugar puckers in this
right-handed parallel-stranded G-quadruplex structure. The
four G.G.G.G tetrad planes stack on each other with minimal
overlap of adjacent guanine base planes within individual
strands. The thymine residues are under-defined in the
solution structure of the d(T-T-G-G-G-G-T) G-quadruplex and
sample amongst multiple conformations in solution.