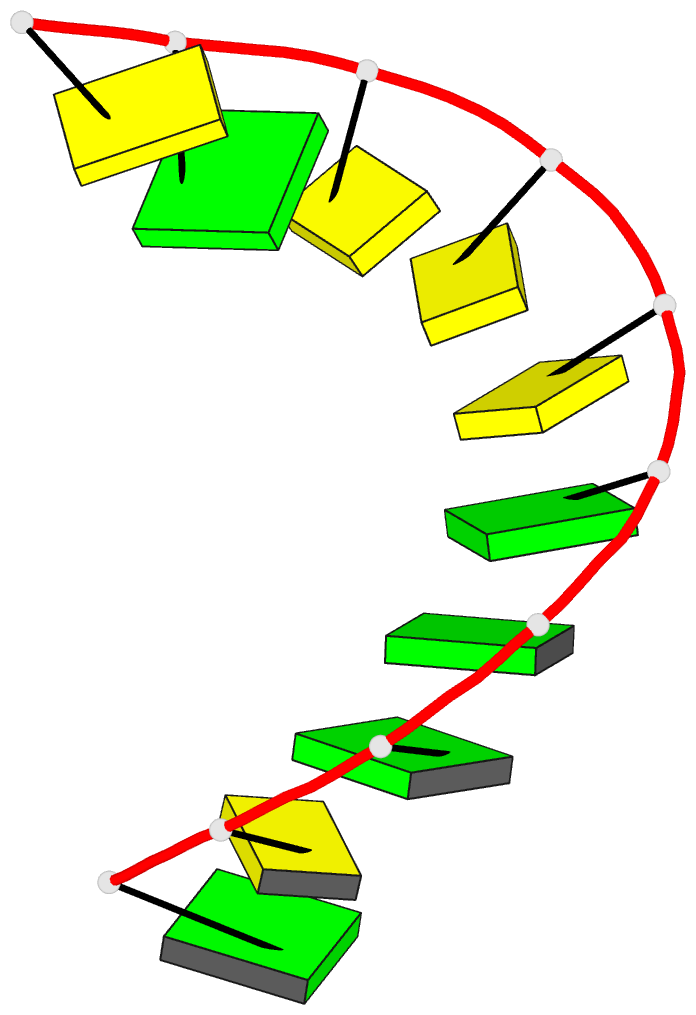
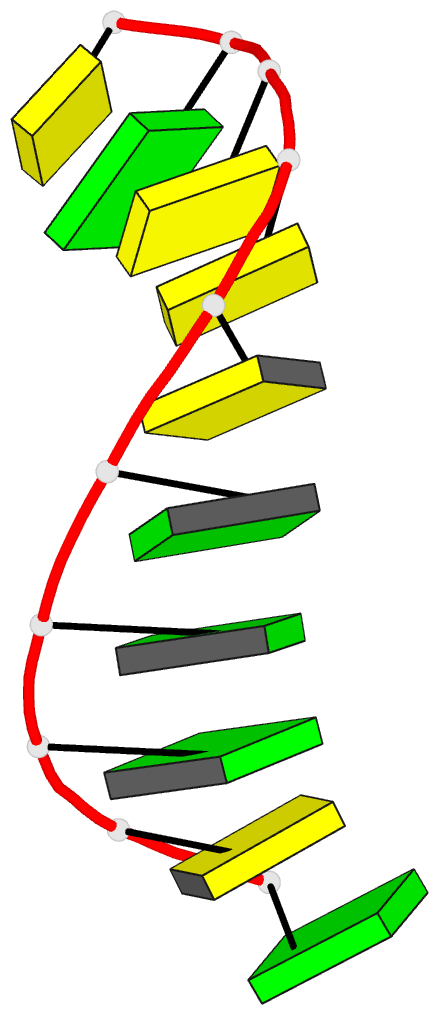
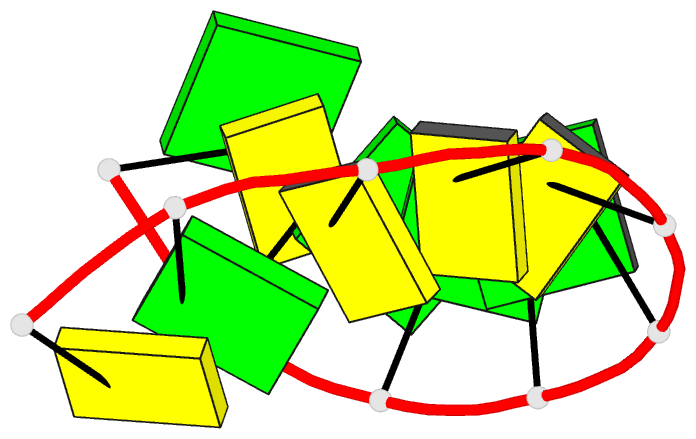
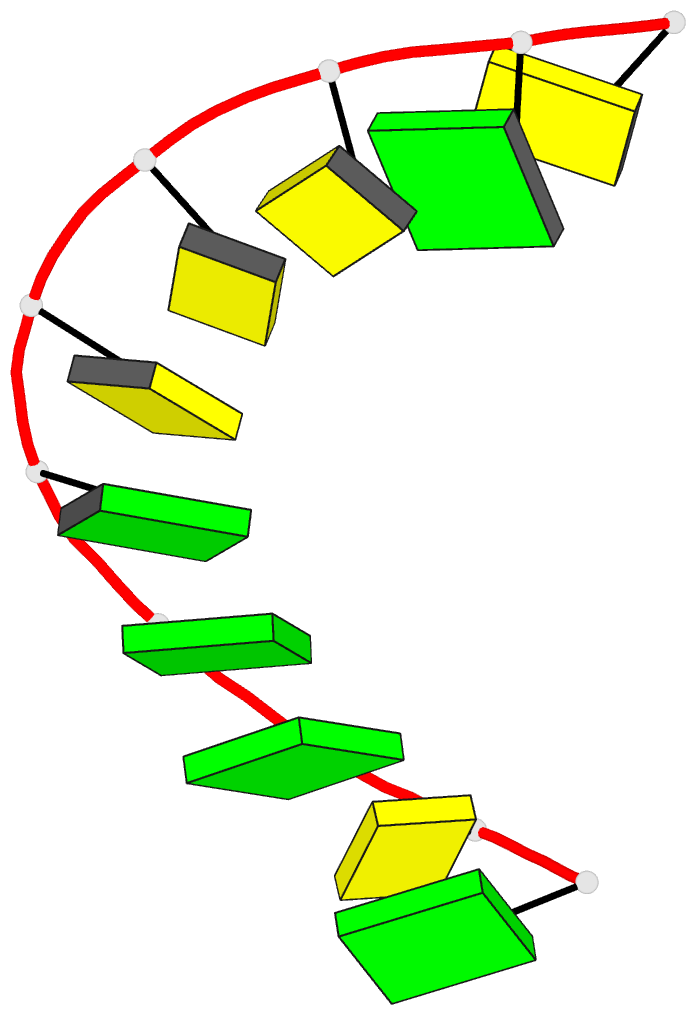
Summary information and primary citation
- PDB-id
-
138d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- X-ray (1.8 Å)
- Summary
- A-DNA decamer d(gcgggcccgc)-hexagonal crystal form
- Reference
-
Ramakrishnan B, Sundaralingam M (1993): "Evidence
for crystal environment dominating base sequence effects
on DNA conformation: crystal structures of the
orthorhombic and hexagonal polymorphs of the A-DNA
decamer d(GCGGGCCCGC) and comparison with their
isomorphous crystal structures."
Biochemistry, 32, 11458-11468.
doi: 10.1021/bi00093a025.
- Abstract
- We have determined the structure of the A-DNA decamer
d(GCGGGCCCGC) in two crystal forms, orthorhombic and
hexagonal, at 1.7- and 1.8-A resolution, respectively. In
the orthorhombic form, the fifth guanine residue has nearly
trans-trans conformations for the alpha-gamma backbone
torsions, as in the isomorphous orthorhombic structure
d(CCCGGCCGGG) [Ramakrishnan, B., & Sundaralingam, M. (1993)
J. Mol. Biol. 231, 431-444]. However, in the hexagonal
form, the eighth cytosine residue adopts the trans-trans
conformations for the backbone alpha-gamma torsions, as in
the isomorphous hexagonal structure d(ACCGGCCGGT)
[Frederick, C. A., Quigley, G. J., Teng, M.-K., Coll, M.,
van der Marel, G. A., van Boom, J. H., Rich, A., & Wang, A.
H.-J. (1989) Eur. J. Biochem. 181, 295-307]. Even though
the average helix and base-pair parameters are nearly the
same in the two polymorphous crystal forms having the same
sequence, many of the base-dependent local helix parameters
are quite different. However, in the isomorphous crystal
forms, in spite of the differing base sequences, the local
helix and base-pair parameters of the duplexes are nearly
the same. This indicates that, in crystals, the local
conformation of a DNA structure is affected severely by the
crystal packing environment rather than by the base
sequence.