Summary information and primary citation
- PDB-id
-
114d;
SNAP-derived features in text and
JSON formats
- Class
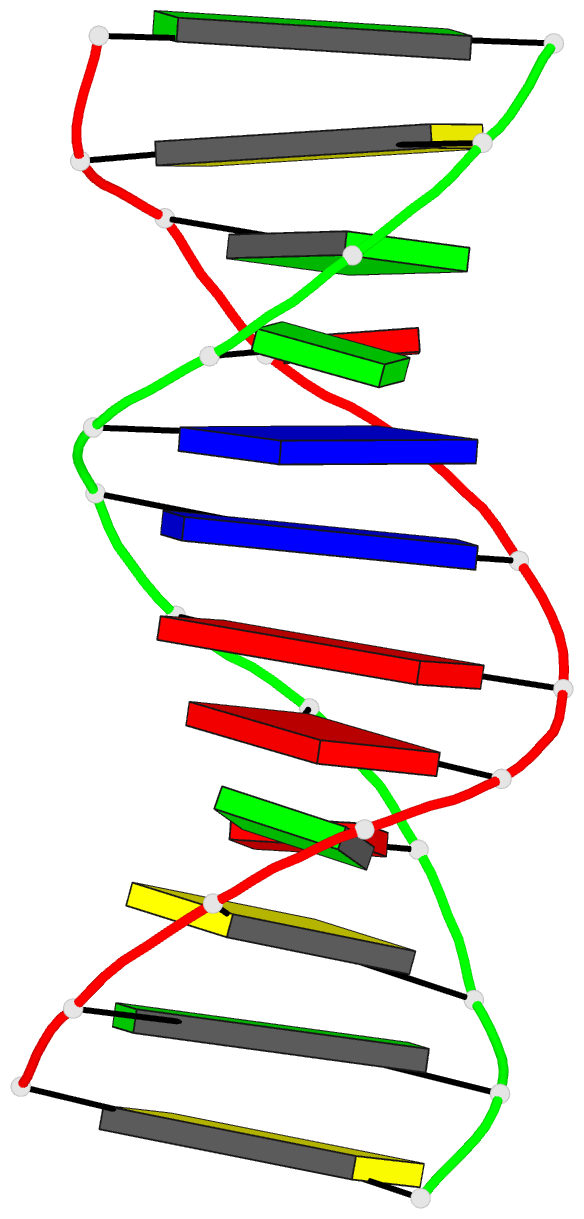
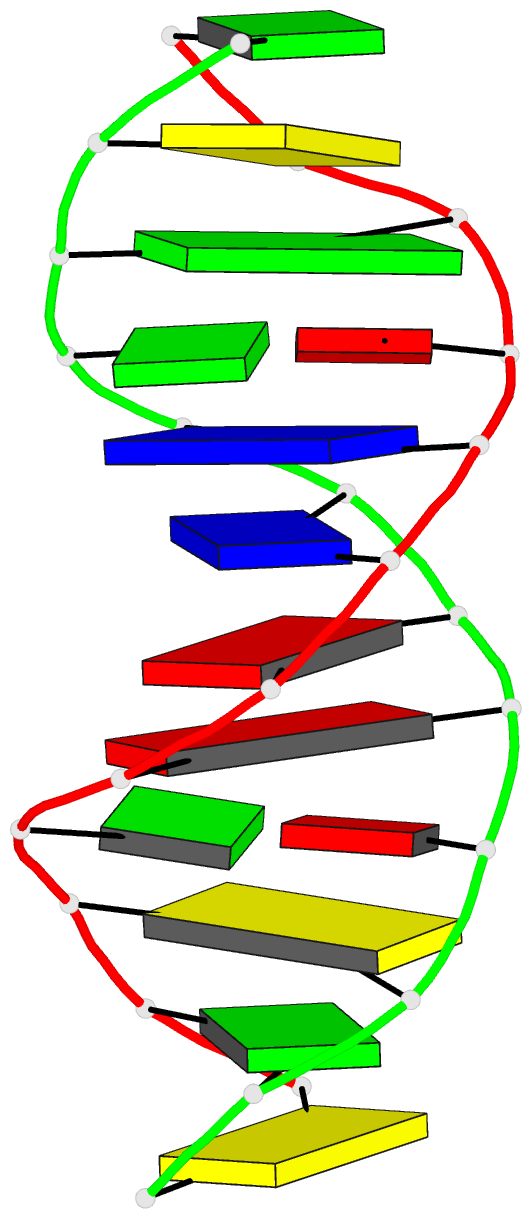
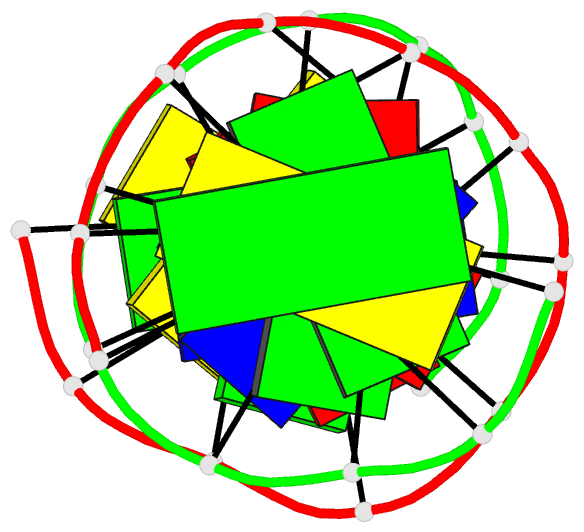
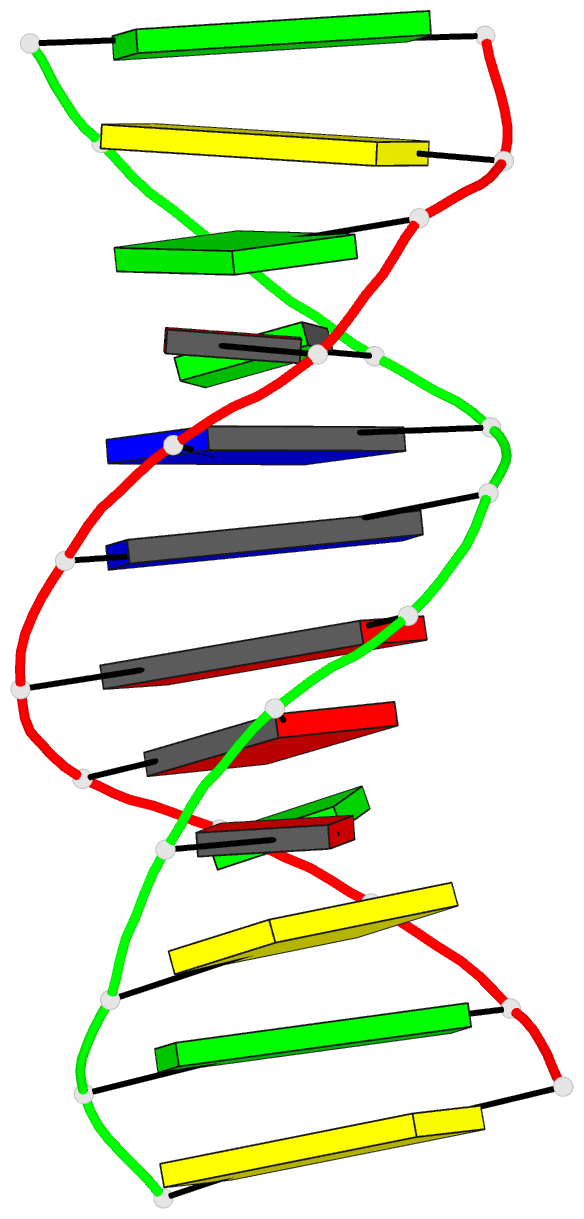
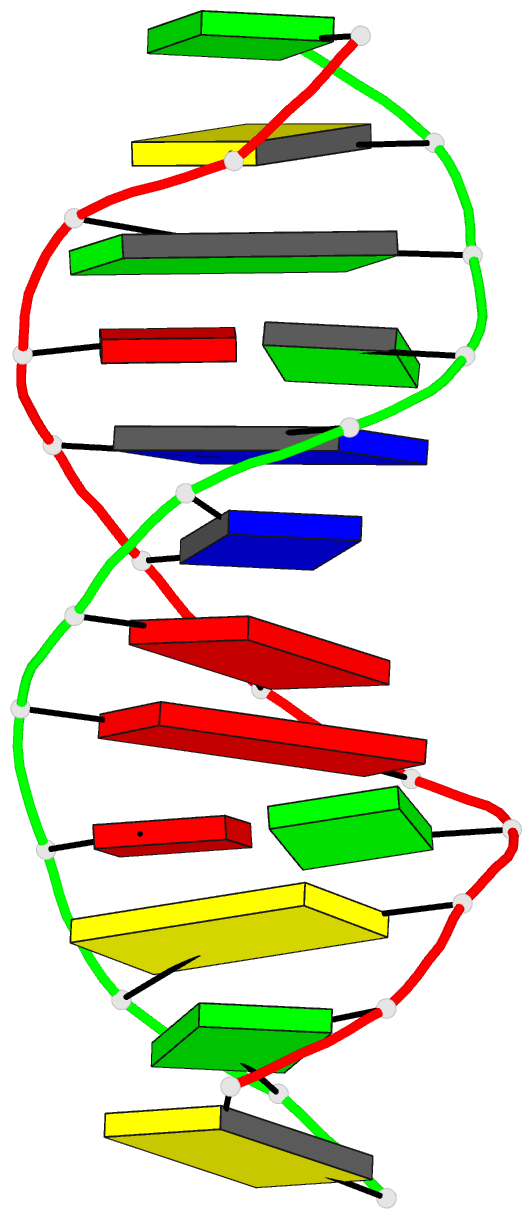
- DNA
- Method
- X-ray (2.5 Å)
- Summary
- Inosine-adenine base pairs in a b-DNA duplex
- Reference
-
Corfield PW, Hunter WN, Brown T, Robinson P, Kennard O
(1987): "Inosine.adenine
base pairs in a B-DNA duplex." Nucleic Acids
Res., 15, 7935-7949. doi: 10.1093/nar/15.19.7935.
- Abstract
- The structure of the synthetic deoxydodecamer
d(C-G-C-I-A-A-T-T-A-G-C-G) has been determined by single
crystal X-ray diffraction techniques at 2.5A resolution.
The refinement converged with a crystallographic residual,
R = 0.19 and the location of 64 solvent molecules. The
sequence crystallises as a B-DNA helix with 10 Watson-Crick
base-pairs (4 A.T. and 6 G.C) and 2 inosine.adenine (I.A)
pairs. The present work shows that in the purine.purine
base-pairs the adenine adopts syn orientation with respect
to the furanose moiety while the inosine is in the trans
(anti) orientation. Two hydrogen bonds link the I.A.
base-pair, one between N-1(I) and N-7(A), the other between
O-6(I) and N-6(A). This bulky purine.purine base-pair is
incorporated in the double helix at two positions with
little distortion of either local or global conformation.
The pairing observed in this study is presented as a model
for I.A base-pairs in RNA codon-anticodon interactions and
may help explain the thermodynamic stability of inosine
containing base-pairs. Conformational parameters and base
stacking interactions are presented and where appropriate
compared with those of the native compound,
d(C-G-C-G-A-A-T-T-C-G-C-G) and with other studies of
oligonucleotides containing purine.purine base-pairs.