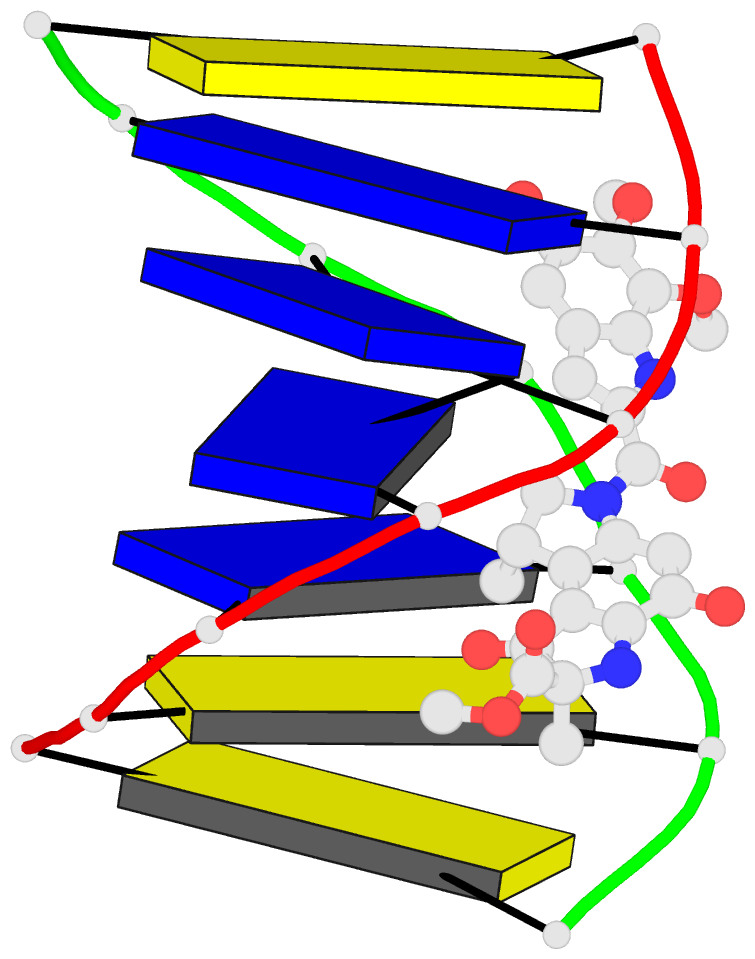
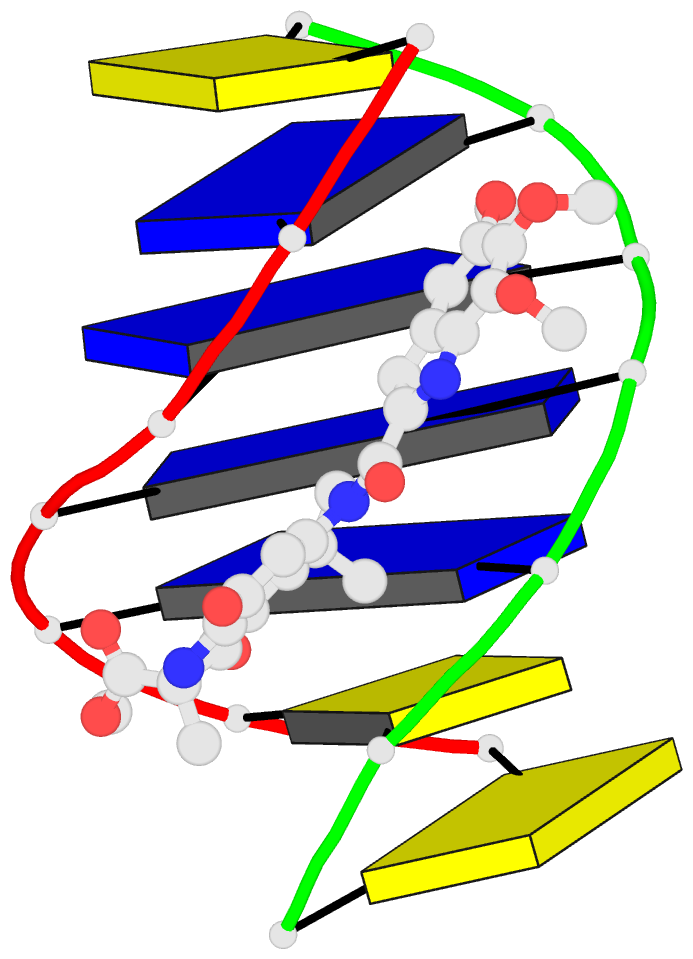
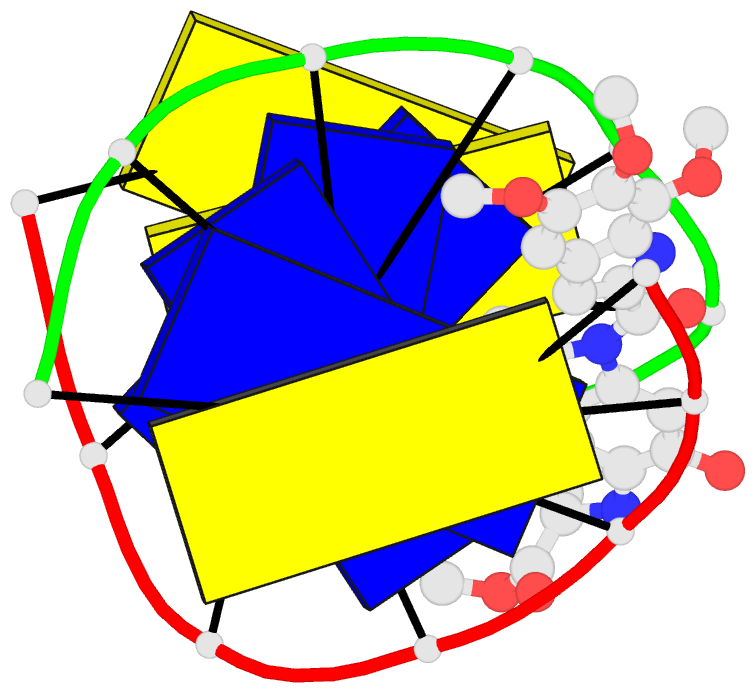
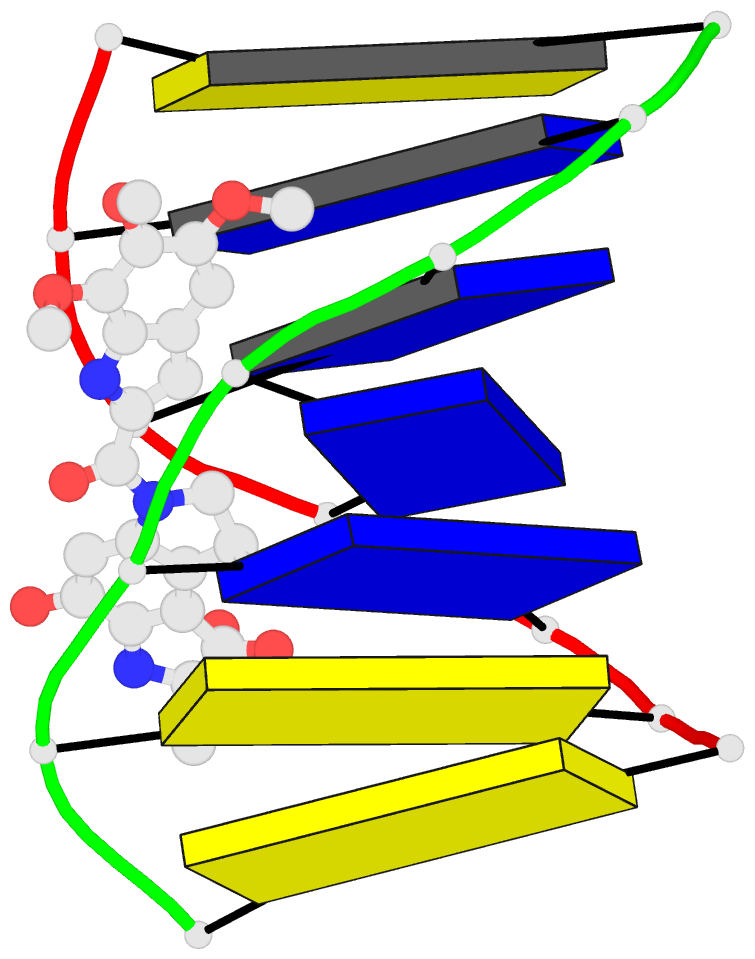
Summary information and primary citation
- PDB-id
-
107d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- NMR
- Summary
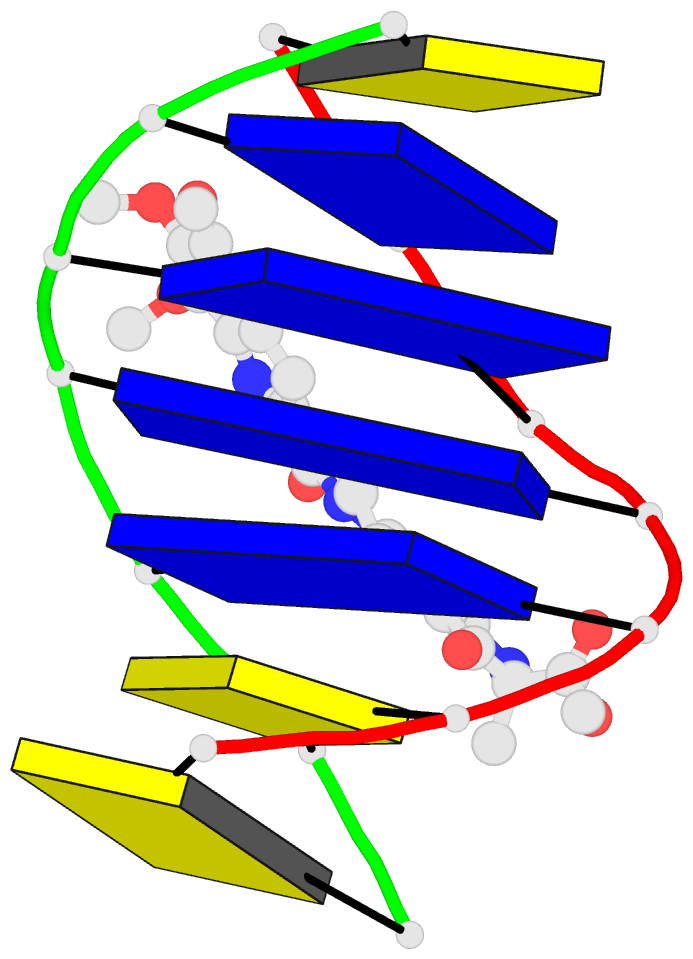
- Solution structure of the covalent duocarmycin a-DNA
duplex complex
- Reference
-
Lin CH, Patel DJ (1995): "Solution
structure of the covalent duocarmycin A-DNA duplex
complex." J.Mol.Biol., 248,
162-179. doi: 10.1006/jmbi.1995.0209.
- Abstract
- Duocarmycin A is an antitumour antibiotic that binds
covalently to the minor groove N-3 position of adenine with
sequence specificity for the 3'-adenine in a d(A-A-A-A)
tract in duplex DNA. The adenine ring becomes protonated on
duocarmycin adduct formation resulting in charge
delocalization over the purine ring system. We report on
the solution structure of duocarmycin A bound site
specifically to A12 (designated *A12+) in the sequence
context d(T3-T4-T5-T6).d(A9-A10-A11-*A12+) within a hairpin
duplex. The solution structure was solved based on a
combined NMR-molecular dynamics study including NOE based
intensity refinement. The A and B-rings of duocarmycin are
positioned deep within the walls of the minor groove with
the B-ring (which is furthest from the covalent linkage
site) directed towards the 5'-end of the modified strand.
Duocarmycin adopts an extended conformation and is aligned
at approximately 45 degrees to the helix axis with its
non-polar concave edges interacting with the floor of the
minor groove while its polar edges are sandwiched within
the walls of the minor groove. The T3.*A12+ modification
site pair forms a weak central Watson-Crick hydrogen bond
in contrast to all A.T and G.C pairs, which align through
standard Watson-Crick pairing in the complex. The helical
parameters are consistent with a minimally perturbed
right-handed duplex in the complex with minor groove width
and x-displacement parameters indicative of a B-form helix.
A striking feature of the complex is the positioning of
duocarmycin A within the walls of the minor groove
resulting in upfield shifts of the minor groove sugar
protons, as well as backbone proton and phosphorus
resonances in the DNA segment spanning the binding
site.